
Part II Plant Sciences Module L1
Reading list for the Synthetic Biology part of the Plant Sciences Part II L1 module
See titles of recommended review articles below - click to download as PDFs
A good starting point:
Synthetic biology: Applications come of age. Khalil, A. S., & Collins, J. J. Nature Reviews. Genetics, 11:367-79 (2010).
Included poster that describes Synthetic Biology applications
Additional reading

Lecture 1: Genetic modification in agriculture and the advent of Synthetic Biology
Genetically engineered plants and foods: A scientist's analysis of the issues (part I). Lemaux, P. G. Annual Review of Plant Biology, 59:771-812 (2008).
Engineering for biofuels: Exploiting innate microbial capacity or importing biosynthetic potential? Alper, H., & Stephanopoulos, G. Nature Reviews. Microbiology, 7:715-23 (2009).
Engineering for biofuels: Exploiting innate microbial capacity or importing biosynthetic potential? Alper, H., & Stephanopoulos, G. Nature Reviews. Microbiology, 7:715-23 (2009).
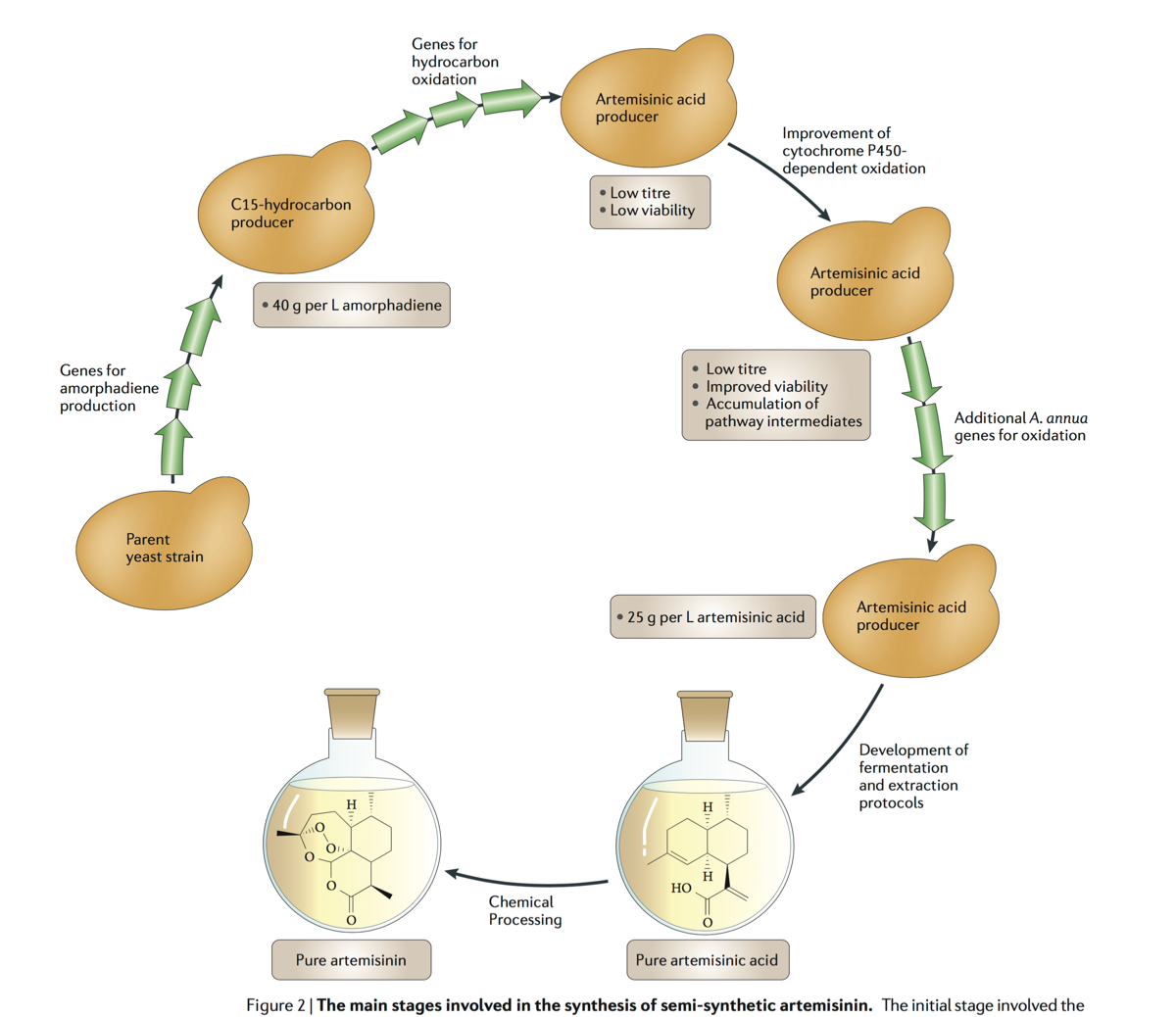
Lecture 2: Genetic circuits and genome scale DNA engineering
Creation of a bacterial cell controlled by a chemically synthesized genome. Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R. Y., Algire, M. A., . . . Venter, J. C. Science (New York, N.Y.), 329:52-6 (2010).
Foundations for engineering biology. Endy, D. Nature, 438:449-53 (2005).
Refinement and standardization of synthetic biological parts and devices. Canton, B., Labno, A., & Endy, D. Nature Biotechnology, 26:787-93 (2008).
The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. YouYou Tu. Nature Medicine 17:1217-1220 (2011).
Foundations for engineering biology. Endy, D. Nature, 438:449-53 (2005).
Refinement and standardization of synthetic biological parts and devices. Canton, B., Labno, A., & Endy, D. Nature Biotechnology, 26:787-93 (2008).
The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. YouYou Tu. Nature Medicine 17:1217-1220 (2011).
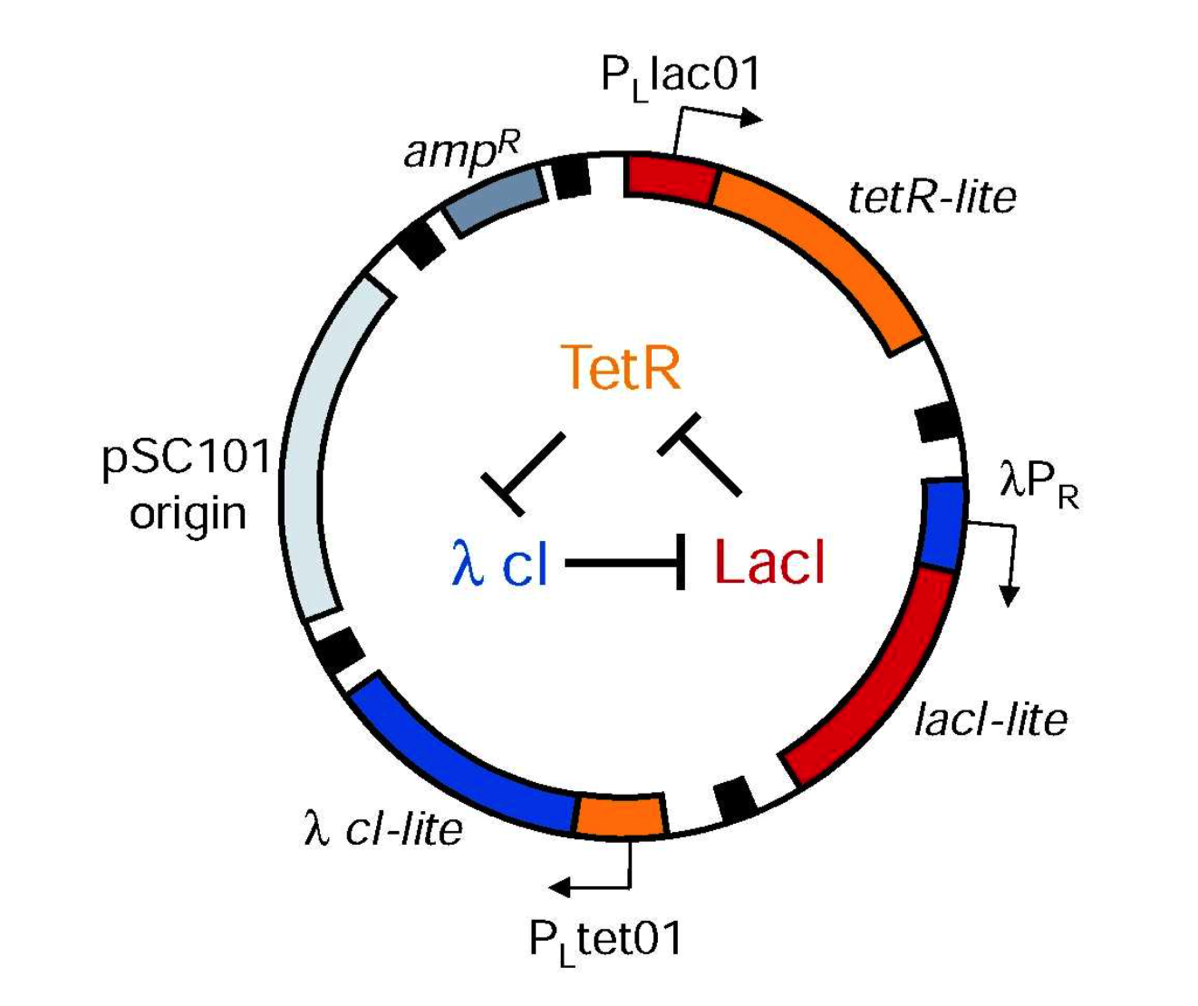
Lecture 3: Engineered logic and the control of gene expression
Designing customized cell signalling circuits. Lim, W. A. Nature Reviews. Molecular Cell Biology, 11:393-403 (2010).
Reconstruction of genetic circuits. Sprinzak, D., & Elowitz, M. B. Nature, 438:443-8 (2005).
Reconstruction of genetic circuits. Sprinzak, D., & Elowitz, M. B. Nature, 438:443-8 (2005).
Lecture 4: Self-organisation and reprogramming of multicellular systems
A synchronized quorum of genetic clocks. Danino, T., Mondragón-Palomino, O., Tsimring, L., & Hasty, J. Nature, 463:326-30 (2010).
Extract from a special Synthetic Biology edition of Science (2nd September 2011)
The Allure of Synthetic Biology; The Life Hacker; Algae’s Second Try; A Lab of Their Own; Visions of Synthetic Biology
REVIEWS
Synthetic Biology: Integrated Gene Circuits, N. Nandagopal and M. B. Elowitz
Synthetic Biology Moving into the Clinic, W. C. Ruder et al.
PERSPECTIVE
Bottom-Up Synthetic Biology: Engineering in a Tinkerer’s World P. Schwille
POLICY FORUM
Synthetic Biology: Regulating Industry Uses of New Biotechnologies, B. Erickson et al.
Download as PDF
The Allure of Synthetic Biology; The Life Hacker; Algae’s Second Try; A Lab of Their Own; Visions of Synthetic Biology
REVIEWS
Synthetic Biology: Integrated Gene Circuits, N. Nandagopal and M. B. Elowitz
Synthetic Biology Moving into the Clinic, W. C. Ruder et al.
PERSPECTIVE
Bottom-Up Synthetic Biology: Engineering in a Tinkerer’s World P. Schwille
POLICY FORUM
Synthetic Biology: Regulating Industry Uses of New Biotechnologies, B. Erickson et al.
Download as PDF
DNA engineering
Engineered gene circuits.
Abstract: A central focus of postgenomic research will be to understand how cellular phenomena arise from the connectivity of genes and proteins. This connectivity generates molecular network diagrams that resemble complex electrical circuits, and a systematic understanding will require the development of a mathematical framework for describing the circuitry. From an engineering perspective, the natural path towards such a framework is the construction and analysis of the underlying submodules that constitute the network. Recent experimental advances in both sequencing and genetic engineering have made this approach feasible through the design and implementation of synthetic gene networks amenable to mathematical modelling and quantitative analysis. These developments have signalled the emergence of a gene circuit discipline, which provides a framework for predicting and evaluating the dynamics of cellular processes. Synthetic gene networks will also lead to new logical forms of cellular control, which could have important applications in functional genomics, nanotechnology, and gene and cell therapy.
Hasty J, McMillen D, Collins JJ. Nature. 420(6912):224-30, (2002).
Reconstruction of genetic circuits.
Abstract: The complex genetic circuits found in cells are ordinarily studied by analysis of genetic and biochemical perturbations. The inherent modularity of biological components like genes and proteins enables a complementary approach: one can construct and analyse synthetic genetic circuits based on their natural counterparts. Such synthetic circuits can be used as simple in vivo models to explore the relation between the structure and function of a genetic circuit. Here we describe recent progress in this area of synthetic biology, highlighting newly developed genetic components and biological lessons learned from this approach.
Sprinzak D , Elowitz MB. Nature. 438(7067):443-8, (2005)
Foundations for engineering biology.
Abstract: Engineered biological systems have been used to manipulate information, construct materials, process chemicals, produce energy, provide food, and help maintain or enhance human health and our environment. Unfortunately, our ability to quickly and reliably engineer biological systems that behave as expected remains quite limited. Foundational technologies that make routine the engineering of biology are needed. Vibrant, open research communities and strategic leadership are necessary to ensure that the development and application of biological technologies remains overwhelmingly constructive.
Endy D. Nature. 438(7067):449-53 (2005)
Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome.
Abstract: We have synthesized a 582,970-base pair Mycoplasma genitalium genome. This synthetic genome, named M. genitalium JCVI-1.0, contains all the genes of wild-type M. genitalium G37 except MG408, which was disrupted by an antibiotic marker to block pathogenicity and to allow for selection. To identify the genome as synthetic, we inserted "watermarks" at intergenic sites known to tolerate transposon insertions. Overlapping "cassettes" of 5 to 7 kilobases (kb), assembled from chemically synthesized oligonucleotides, were joined by in vitro recombination to produce intermediate assemblies of approximately 24 kb, 72 kb ("1/8 genome"), and 144 kb ("1/4 genome"), which were all cloned as bacterial artificial chromosomes in Escherichia coli. Most of these intermediate clones were sequenced, and clones of all four 1/4 genomes with the correct sequence were identified. The complete synthetic genome was assembled by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae, then isolated and sequenced. A clone with the correct sequence was identified. The methods described here will be generally useful for constructing large DNA molecules from chemically synthesized pieces and also from combinations of natural and synthetic DNA segments.
Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA 3rd, Smith HO. Science. 319(5867):1215-20, (2008).
Creation of a bacterial cell controlled by a chemically synthesized genome.
Abstract: We report the design, synthesis, and assembly of the 1.08-mega-base pair Mycoplasma mycoides JCVI-syn1.0 genome starting from digitized genome sequence information and its transplantation into a M. capricolum recipient cell to create new M. mycoides cells that are controlled only by the synthetic chromosome. The only DNA in the cells is the designed synthetic DNA sequence, including "watermark" sequences and other designed gene deletions and polymorphisms, and mutations acquired during the building process. The new cells have expected phenotypic properties and are capable of continuous self-replication.
Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA 3rd, Smith HO, Venter JC. Science. 329(5987):52-6. (2010)
Engineered gene circuits.
Abstract: A central focus of postgenomic research will be to understand how cellular phenomena arise from the connectivity of genes and proteins. This connectivity generates molecular network diagrams that resemble complex electrical circuits, and a systematic understanding will require the development of a mathematical framework for describing the circuitry. From an engineering perspective, the natural path towards such a framework is the construction and analysis of the underlying submodules that constitute the network. Recent experimental advances in both sequencing and genetic engineering have made this approach feasible through the design and implementation of synthetic gene networks amenable to mathematical modelling and quantitative analysis. These developments have signalled the emergence of a gene circuit discipline, which provides a framework for predicting and evaluating the dynamics of cellular processes. Synthetic gene networks will also lead to new logical forms of cellular control, which could have important applications in functional genomics, nanotechnology, and gene and cell therapy.
Hasty J, McMillen D, Collins JJ. Nature. 420(6912):224-30, (2002).
Reconstruction of genetic circuits.
Abstract: The complex genetic circuits found in cells are ordinarily studied by analysis of genetic and biochemical perturbations. The inherent modularity of biological components like genes and proteins enables a complementary approach: one can construct and analyse synthetic genetic circuits based on their natural counterparts. Such synthetic circuits can be used as simple in vivo models to explore the relation between the structure and function of a genetic circuit. Here we describe recent progress in this area of synthetic biology, highlighting newly developed genetic components and biological lessons learned from this approach.
Sprinzak D , Elowitz MB. Nature. 438(7067):443-8, (2005)
Foundations for engineering biology.
Abstract: Engineered biological systems have been used to manipulate information, construct materials, process chemicals, produce energy, provide food, and help maintain or enhance human health and our environment. Unfortunately, our ability to quickly and reliably engineer biological systems that behave as expected remains quite limited. Foundational technologies that make routine the engineering of biology are needed. Vibrant, open research communities and strategic leadership are necessary to ensure that the development and application of biological technologies remains overwhelmingly constructive.
Endy D. Nature. 438(7067):449-53 (2005)
Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome.
Abstract: We have synthesized a 582,970-base pair Mycoplasma genitalium genome. This synthetic genome, named M. genitalium JCVI-1.0, contains all the genes of wild-type M. genitalium G37 except MG408, which was disrupted by an antibiotic marker to block pathogenicity and to allow for selection. To identify the genome as synthetic, we inserted "watermarks" at intergenic sites known to tolerate transposon insertions. Overlapping "cassettes" of 5 to 7 kilobases (kb), assembled from chemically synthesized oligonucleotides, were joined by in vitro recombination to produce intermediate assemblies of approximately 24 kb, 72 kb ("1/8 genome"), and 144 kb ("1/4 genome"), which were all cloned as bacterial artificial chromosomes in Escherichia coli. Most of these intermediate clones were sequenced, and clones of all four 1/4 genomes with the correct sequence were identified. The complete synthetic genome was assembled by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae, then isolated and sequenced. A clone with the correct sequence was identified. The methods described here will be generally useful for constructing large DNA molecules from chemically synthesized pieces and also from combinations of natural and synthetic DNA segments.
Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA 3rd, Smith HO. Science. 319(5867):1215-20, (2008).
Creation of a bacterial cell controlled by a chemically synthesized genome.
Abstract: We report the design, synthesis, and assembly of the 1.08-mega-base pair Mycoplasma mycoides JCVI-syn1.0 genome starting from digitized genome sequence information and its transplantation into a M. capricolum recipient cell to create new M. mycoides cells that are controlled only by the synthetic chromosome. The only DNA in the cells is the designed synthetic DNA sequence, including "watermark" sequences and other designed gene deletions and polymorphisms, and mutations acquired during the building process. The new cells have expected phenotypic properties and are capable of continuous self-replication.
Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA 3rd, Smith HO, Venter JC. Science. 329(5987):52-6. (2010)
Circuit design
Construction of a genetic toggle switch in Escherichia coli.
Abstract: It has been proposed' that gene-regulatory circuits with virtually any desired property can be constructed from networks of simple regulatory elements. These properties, which include multistability and oscillations, have been found in specialized gene circuits such as the bacteriophage lambda switch and the Cyanobacteria circadian oscillator. However, these behaviours have not been demonstrated in networks of non-specialized regulatory components. Here we present the construction of a genetic toggle switch-a synthetic, bistable gene-regulatory network-in Escherichia coli and provide a simple theory that predicts the conditions necessary for bistability. The toggle is constructed from any two repressible promoters arranged in a mutually inhibitory network. It is flipped between stable states using transient chemical or thermal induction and exhibits a nearly ideal switching threshold. As a practical device, the toggle switch forms a synthetic, addressable cellular memory unit and has implications forbiotechnology, biocomputing and gene therapy.
Gardner TS, Cantor CR, Collins JJ. Nature. 403(6767):339-42 (2000)
A synthetic oscillatory network of transcriptional regulators.
Abstract: Networks of interacting biomolecules carry out many essential functions in living cells, but the 'design principles' underlying the functioning of such intracellular networks remain poorly understood, despite intensive efforts including quantitative analysis of relatively simple systems. Here we present a complementary approach to this problem: the design and construction of a synthetic network to implement a particular function. We used three transcriptional repressor systems that are not part of any natural biological clock to build an oscillating network, termed the repressilator, in Escherichia coli. The network periodically induces the synthesis of green fluorescent protein as a readout of its state in individual cells. The resulting oscillations, with typical periods of hours, are slower than the cell-division cycle, so the state of the oscillator has to be transmitted from generation to generation. This artificial clock displays noisy behaviour, possibly because of stochastic fluctuations of its components. Such 'rational network design may lead both to the engineering of new cellular behaviours and to an improved understanding of naturally occurring networks.
Elowitz MB, Leibler S. Nature. 403(6767):335-8. (2000)
A synthetic multicellular system for programmed pattern formation.
Abstract: Pattern formation is a hallmark of coordinated cell behaviour in both single and multicellular organisms. It typically involves cell-cell communication and intracellular signal processing. Here we show a synthetic multicellular system in which genetically engineered 'receiver' cells are programmed to form ring-like patterns of differentiation based on chemical gradients of an acyl-homoserine lactone (AHL) signal that is synthesized by 'sender' cells. In receiver cells, 'band-detect' gene networks respond to user-defined ranges of AHL concentrations. By fusing different fluorescent proteins as outputs of network variants, an initially undifferentiated 'lawn' of receivers is engineered to form a bullseye pattern around a sender colony. Other patterns, such as ellipses and clovers, are achieved by placing senders in different configurations. Experimental and theoretical analyses reveal which kinetic parameters most significantly affect ring development over time. Construction and study of such synthetic multicellular systems can improve our quantitative understanding of naturally occurring developmental processes and may foster applications in tissue engineering, biomaterial fabrication and biosensing.
Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. Nature. 434(7037):1130-4, (2005).
Production of the antimalarial drug precursor artemisinic acid in engineered yeast.
Abstract: Malaria is a global health problem that threatens 300-500 million people and kills more than one million people annually. Disease control is hampered by the occurrence of multi-drug-resistant strains of the malaria parasite Plasmodium falciparum. Synthetic antimalarial drugs and malarial vaccines are currently being developed, but their efficacy against malaria awaits rigorous clinical testing. Artemisinin, a sesquiterpene lactone endoperoxide extracted from Artemisia annua L (family Asteraceae; commonly known as sweet wormwood), is highly effective against multi-drug-resistant Plasmodium spp., but is in short supply and unaffordable to most malaria sufferers. Although total synthesis of artemisinin is difficult and costly, the semi-synthesis of artemisinin or any derivative from microbially sourced artemisinic acid, its immediate precursor, could be a cost-effective, environmentally friendly, high-quality and reliable source of artemisinin. Here we report the engineering of Saccharomyces cerevisiae to produce high titres (up to 100 mg l-1) of artemisinic acid using an engineered mevalonate pathway, amorphadiene synthase, and a novel cytochrome P450 monooxygenase (CYP71AV1) from A. annua that performs a three-step oxidation of amorpha-4,11-diene to artemisinic acid. The synthesized artemisinic acid is transported out and retained on the outside of the engineered yeast, meaning that a simple and inexpensive purification process can be used to obtain the desired product. Although the engineered yeast is already capable of producing artemisinic acid at a significantly higher specific productivity than A. annua, yield optimization and industrial scale-up will be required to raise artemisinic acid production to a level high enough to reduce artemisinin combination therapies to significantly below their current prices.
Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 440(7086):940-3, (2006).
Construction of a genetic toggle switch in Escherichia coli.
Abstract: It has been proposed' that gene-regulatory circuits with virtually any desired property can be constructed from networks of simple regulatory elements. These properties, which include multistability and oscillations, have been found in specialized gene circuits such as the bacteriophage lambda switch and the Cyanobacteria circadian oscillator. However, these behaviours have not been demonstrated in networks of non-specialized regulatory components. Here we present the construction of a genetic toggle switch-a synthetic, bistable gene-regulatory network-in Escherichia coli and provide a simple theory that predicts the conditions necessary for bistability. The toggle is constructed from any two repressible promoters arranged in a mutually inhibitory network. It is flipped between stable states using transient chemical or thermal induction and exhibits a nearly ideal switching threshold. As a practical device, the toggle switch forms a synthetic, addressable cellular memory unit and has implications forbiotechnology, biocomputing and gene therapy.
Gardner TS, Cantor CR, Collins JJ. Nature. 403(6767):339-42 (2000)
A synthetic oscillatory network of transcriptional regulators.
Abstract: Networks of interacting biomolecules carry out many essential functions in living cells, but the 'design principles' underlying the functioning of such intracellular networks remain poorly understood, despite intensive efforts including quantitative analysis of relatively simple systems. Here we present a complementary approach to this problem: the design and construction of a synthetic network to implement a particular function. We used three transcriptional repressor systems that are not part of any natural biological clock to build an oscillating network, termed the repressilator, in Escherichia coli. The network periodically induces the synthesis of green fluorescent protein as a readout of its state in individual cells. The resulting oscillations, with typical periods of hours, are slower than the cell-division cycle, so the state of the oscillator has to be transmitted from generation to generation. This artificial clock displays noisy behaviour, possibly because of stochastic fluctuations of its components. Such 'rational network design may lead both to the engineering of new cellular behaviours and to an improved understanding of naturally occurring networks.
Elowitz MB, Leibler S. Nature. 403(6767):335-8. (2000)
A synthetic multicellular system for programmed pattern formation.
Abstract: Pattern formation is a hallmark of coordinated cell behaviour in both single and multicellular organisms. It typically involves cell-cell communication and intracellular signal processing. Here we show a synthetic multicellular system in which genetically engineered 'receiver' cells are programmed to form ring-like patterns of differentiation based on chemical gradients of an acyl-homoserine lactone (AHL) signal that is synthesized by 'sender' cells. In receiver cells, 'band-detect' gene networks respond to user-defined ranges of AHL concentrations. By fusing different fluorescent proteins as outputs of network variants, an initially undifferentiated 'lawn' of receivers is engineered to form a bullseye pattern around a sender colony. Other patterns, such as ellipses and clovers, are achieved by placing senders in different configurations. Experimental and theoretical analyses reveal which kinetic parameters most significantly affect ring development over time. Construction and study of such synthetic multicellular systems can improve our quantitative understanding of naturally occurring developmental processes and may foster applications in tissue engineering, biomaterial fabrication and biosensing.
Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. Nature. 434(7037):1130-4, (2005).
Production of the antimalarial drug precursor artemisinic acid in engineered yeast.
Abstract: Malaria is a global health problem that threatens 300-500 million people and kills more than one million people annually. Disease control is hampered by the occurrence of multi-drug-resistant strains of the malaria parasite Plasmodium falciparum. Synthetic antimalarial drugs and malarial vaccines are currently being developed, but their efficacy against malaria awaits rigorous clinical testing. Artemisinin, a sesquiterpene lactone endoperoxide extracted from Artemisia annua L (family Asteraceae; commonly known as sweet wormwood), is highly effective against multi-drug-resistant Plasmodium spp., but is in short supply and unaffordable to most malaria sufferers. Although total synthesis of artemisinin is difficult and costly, the semi-synthesis of artemisinin or any derivative from microbially sourced artemisinic acid, its immediate precursor, could be a cost-effective, environmentally friendly, high-quality and reliable source of artemisinin. Here we report the engineering of Saccharomyces cerevisiae to produce high titres (up to 100 mg l-1) of artemisinic acid using an engineered mevalonate pathway, amorphadiene synthase, and a novel cytochrome P450 monooxygenase (CYP71AV1) from A. annua that performs a three-step oxidation of amorpha-4,11-diene to artemisinic acid. The synthesized artemisinic acid is transported out and retained on the outside of the engineered yeast, meaning that a simple and inexpensive purification process can be used to obtain the desired product. Although the engineered yeast is already capable of producing artemisinic acid at a significantly higher specific productivity than A. annua, yield optimization and industrial scale-up will be required to raise artemisinic acid production to a level high enough to reduce artemisinin combination therapies to significantly below their current prices.
Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 440(7086):940-3, (2006).
