Microscopy of GFP-expressing plant tissues
The major advantage of GFP is that it is intrinsically fluorescent, and the protein is be directly visible in living tissues where it is being expressed. There are many benefits to working with live samples, and in our experience we get much higher quality images in the absence of fixation. Sample preparation is of two types. For routine short term observation, we simply mount fluorescent protein expressing plant material in water under a coverslip. It is advantageous to counterstain cell walls, e.g. for a few minutes with 10 ug/ml propidium iodide solution. A typical image is shown below.
The major advantage of GFP is that it is intrinsically fluorescent, and the protein is be directly visible in living tissues where it is being expressed. There are many benefits to working with live samples, and in our experience we get much higher quality images in the absence of fixation. Sample preparation is of two types. For routine short term observation, we simply mount fluorescent protein expressing plant material in water under a coverslip. It is advantageous to counterstain cell walls, e.g. for a few minutes with 10 ug/ml propidium iodide solution. A typical image is shown below.
For longer term observations of Arabidopsis roots (greater than a couple of hours) we grow the seedlings in agar-containing sterile culture vessels which have a coverslip base. In these vessels, seeds can germinate on the agar and roots will grow down and then along the surface of the coverslip base - allowing high resolution imaging.
Living plant tissue is a difficult material for high resolution fluorescence imaging. It is aqueous, with layers of refractile cell walls and is typically thick. However, most microscope objectives are designed for use with thin sections mounted in a high refractive index medium. To partially circumvent some of these problems, we use a lens (Nikon PlanApo 60x NA 1.2) corrected for water immersion and the use of coverslips and with a long working distance (nominally 220 microns). This minimises spherical abberation when imaging deep into plant wholemounts. The flat field and high numerical aperture are also beneficial for confocal microscopy.
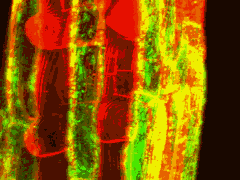
GFP can be excited by the 488nm line of argon and argon-krypton lasers commonly used for confocal microscopy. In addition, chlorophyll autofluorescence or red fluorescent counterstains such as propidium iodide or rhodamine B can be excited by 514nm (argon) or 568nm (argon-krypton) laser light, and the red emission of these fluors is readily separated from the green emission of GFP using dual channel filter sets used for FITC/TRITC imaging. The examples below show GFP and chloroplasts within transformed Arabidopsis mesophyll cells, and a transgenic Arabidopsis root tip which expresses a GAL4-GFP enhancer trap in the endodermis and cortex layers. The root tip has been counterstained with propidium iodide to label the cell walls. There is little overlap of the red and green channels.
Living plant tissue is a difficult material for high resolution fluorescence imaging. It is aqueous, with layers of refractile cell walls and is typically thick. However, most microscope objectives are designed for use with thin sections mounted in a high refractive index medium. To partially circumvent some of these problems, we use a lens (Nikon PlanApo 60x NA 1.2) corrected for water immersion and the use of coverslips and with a long working distance (nominally 220 microns). This minimises spherical abberation when imaging deep into plant wholemounts. The flat field and high numerical aperture are also beneficial for confocal microscopy.
GFP can be excited by the 488nm line of argon and argon-krypton lasers commonly used for confocal microscopy. In addition, chlorophyll autofluorescence or red fluorescent counterstains such as propidium iodide or rhodamine B can be excited by 514nm (argon) or 568nm (argon-krypton) laser light, and the red emission of these fluors is readily separated from the green emission of GFP using dual channel filter sets used for FITC/TRITC imaging. The examples below show GFP and chloroplasts within transformed Arabidopsis mesophyll cells, and a transgenic Arabidopsis root tip which expresses a GAL4-GFP enhancer trap in the endodermis and cortex layers. The root tip has been counterstained with propidium iodide to label the cell walls. There is little overlap of the red and green channels.
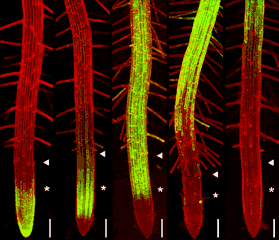
Above left: 35S-mgfp mesophyll cells. Above right: mgfp5-ER Arabidopsis enhancer trap line.
Green channel (488nm excitation, 510-560nm emission) - GFP fluorescence
Red channel (568nm excitation, 580-660nm emission) - chlorophyll & propidium iodide fluorescence
Green channel (488nm excitation, 510-560nm emission) - GFP fluorescence
Red channel (568nm excitation, 580-660nm emission) - chlorophyll & propidium iodide fluorescence
Optical sectioning
The 3D arrangements of plant cells can be observed using either physical or optical sectioning techniques. However the laborious nature of thin sectioning, the problem of obtaining the desired plane of section, and difficulty of obtaining a complete series of section has limited its use somewhat to the skilled and patient. Optical sectioning has many advantages from the point of view of speed and simplicity, and it can allow the direct viewing of living wholemounts. Here, transverse sections can be reconstructed from a series of Z-axis images. Nomarski optics have proved useful for examining details within living roots, but do not provide sufficient contrast and resolution to allow precise 3D reconstruction of cell arrangements throughout the tissue. Confocal laser scanning microscopy allows exclusion of out-of-focus light, and provides substantially improved optical sectioning.
The 3D arrangements of plant cells can be observed using either physical or optical sectioning techniques. However the laborious nature of thin sectioning, the problem of obtaining the desired plane of section, and difficulty of obtaining a complete series of section has limited its use somewhat to the skilled and patient. Optical sectioning has many advantages from the point of view of speed and simplicity, and it can allow the direct viewing of living wholemounts. Here, transverse sections can be reconstructed from a series of Z-axis images. Nomarski optics have proved useful for examining details within living roots, but do not provide sufficient contrast and resolution to allow precise 3D reconstruction of cell arrangements throughout the tissue. Confocal laser scanning microscopy allows exclusion of out-of-focus light, and provides substantially improved optical sectioning.
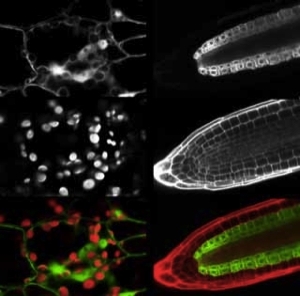
Optical sectioning of a propidium iodide stained root tip of a GAL4-GFP enhancer trap Arabidopsis line (Q2500) is shown at the right. Propidium iodide stains the cell walls of living root cells, and the green fluorescence of GFP can be seen distributed through the pericycle, endodermis and cortex of the root meristem. However, it is still difficult to optically section deep into live tissues due to light scattering and spherical aberration caused by particulate subcellular matter and layers of refractile cell walls.
Instead, the tissues can be fixed, stained and cleared with a high refractive index mountant. The fluorescent labelled plant material can then be observed at high resolution deep into the tissue (>200 microns). Stained and cleared material from the cotyledon tip of a mature Arabidopsis embryo is shown excavated below. The outlines of stained cells are clearly apparent throughout the tissue. We can use this type of data set for 3D segmentation - to reconstruct the precise three dimensional arrangements of developing cells.
Instead, the tissues can be fixed, stained and cleared with a high refractive index mountant. The fluorescent labelled plant material can then be observed at high resolution deep into the tissue (>200 microns). Stained and cleared material from the cotyledon tip of a mature Arabidopsis embryo is shown excavated below. The outlines of stained cells are clearly apparent throughout the tissue. We can use this type of data set for 3D segmentation - to reconstruct the precise three dimensional arrangements of developing cells.



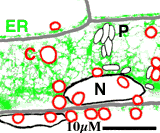
Subcellular dynamics
A transgenic Arabidopsis seedling expressing the mgfp5-ER gene was mounted in water and a small segment of the hypocotyl epidermis was observed using a Biorad MRC-600 laser scanning confocal microscope. The laser light was attenuated by 99% using a neutral density filter, and the confocal aperture was stopped down. Two channel, single scan images were collected at the rate of 1 per 2 seconds for 20 minutes, and transferred to an Apple Macintosh computer. The large data file was then converted to full-colour numbered PICT files using the program PicMerge, and finally converted to a Quicktime movie for analysis and video rate playback.
A transgenic Arabidopsis seedling expressing the mgfp5-ER gene was mounted in water and a small segment of the hypocotyl epidermis was observed using a Biorad MRC-600 laser scanning confocal microscope. The laser light was attenuated by 99% using a neutral density filter, and the confocal aperture was stopped down. Two channel, single scan images were collected at the rate of 1 per 2 seconds for 20 minutes, and transferred to an Apple Macintosh computer. The large data file was then converted to full-colour numbered PICT files using the program PicMerge, and finally converted to a Quicktime movie for analysis and video rate playback.
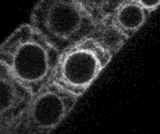
A small section of this movie is shown right as an animated GIF file. Nuclei (N), chloroplasts (C) endoplasmic reticulum (ER), proplastids or leucoplasts (P) are indicated (scale bar = 10 microns)
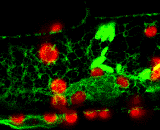
cell division in the root meristem
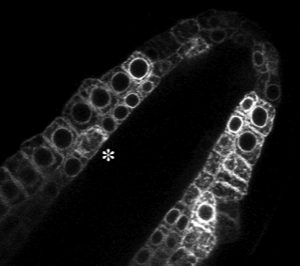
The endoplasmic reticulum localised GFP (mgfp5-ER) provides a useful marker for cell division within the root meristem. Nuclei of the small meristematic cells are clearly highlighted by the perinuclear distribution of GFP in transgenic plants.The animation above shows a small portion of a larger and longer film - from a 6 hour timelapse experiment. A cell is shown caught in the process of mitosis. The division takes approximately 20 minutes. A view of the root meristem from which this animation was taken is shown below. The dividing cell is marked with an asterisk.
The endoplasmic reticulum localised GFP (mgfp5-ER) provides a useful marker for cell division within the root meristem. Nuclei of the small meristematic cells are clearly highlighted by the perinuclear distribution of GFP in transgenic plants.The animation above shows a small portion of a larger and longer film - from a 6 hour timelapse experiment. A cell is shown caught in the process of mitosis. The division takes approximately 20 minutes. A view of the root meristem from which this animation was taken is shown below. The dividing cell is marked with an asterisk.