To help in the design of primers for both PCR and Gibson assembly the University of Cambridge iGEM2010 team have developed a website called Gibthon (http://www.gibthon.org/). A primer pair is needed to inverse PCR everything except mCherry. The second primer pair is required to amplify the super folder GFP gene. Both primers pairs require additional DNA sequences to act as homology drivers and to allow end-linking by Gibson assembly. [Note sequences should avoid the prefix and suffix motifs used for restriction enzyme assembly].

5’-tactagagccaggcatcaaataaaacgaaaggctcagtcg-3’
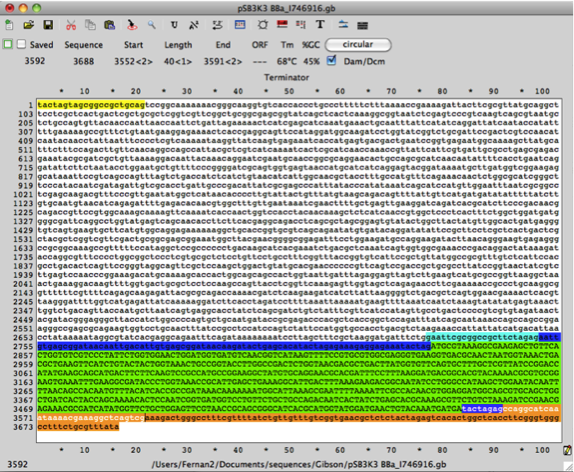
The following DNA sequence has 3’ homology of the super folder GFP gene and the 3’ DNA sequence of pSB3K3 (with promoter and rbs DNA sequences). Copy the DNA sequence:
5’-ATCACGCATGGTATGGATGAACTGTACAAATGATGA-3’
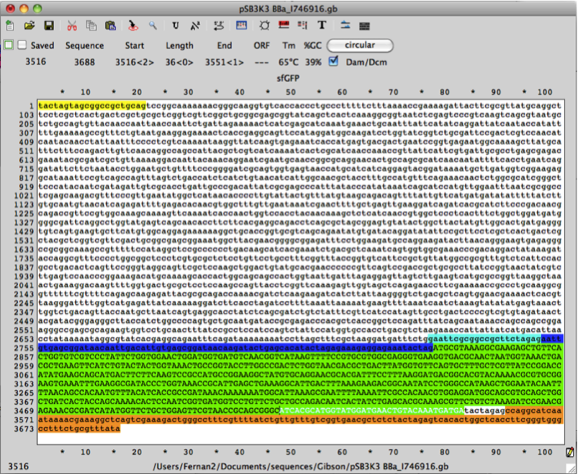
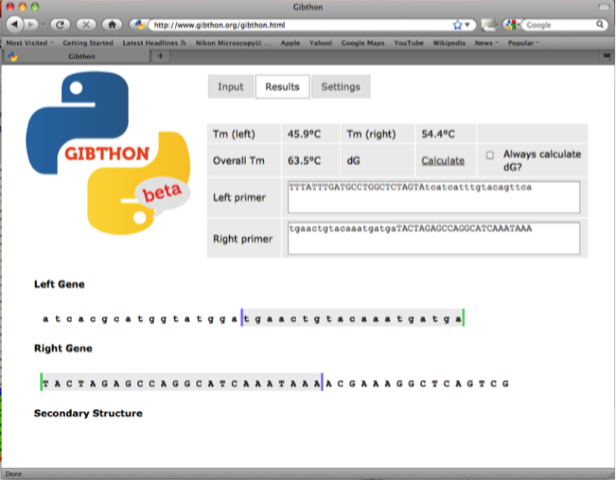
fwd backbone 5’-tgaactgtacaaatgatgaTACTAGAGCCAGGCATCAAATAAA-3’
The left primer is the reverse complement of the right primer and is used as the reverse primer too amplify the super folder GFP gene (with 20 nucleotide homology to the super folder GFP and a 24 nucleotide 5’ overhang)
5’-TTTATTTGATGCCTGGCTCTAGTAtcatcatttgtacagttca-3’
modified slightly to:
rev GFP: 5’-CCTGGCTCTAGTATCAtcatcatttgtacagttcatccatacc-3’
Repeat step 4 to design the second primer pair for PCR and Gibson assembly.
fwd GFP: 5’-TACTAGAGAAAGAGGAGAAATACTAGATGCGTAAAGG-3’
rev backbone: 5’-CATCTAGTATTTCTCCTCTTTCTCTAGTATGTGC-3’
The fwd and rev GFP primers and the fwd and rev plasmid backbone primers are paired for PCR. [Note the primer melting points need to be calculated for the DNA sequence that shares homology with the template DNA to work out the correct annealing temperature for PCR].
