New reagents and techniques for the construction of genetic circuits
High throughput - multiplex - single tube - isothermal - seamless - DNA assembly for everybody!
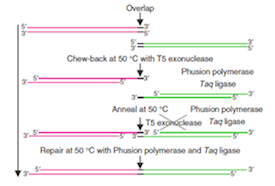
Gibson Assembly is a high-efficiency DNA end-linking technique developed by Daniel Gibson at the JCVI. The technique was invented and perfected as part of the genome assembly efforts at JCVI. The method uses three enzymes to join two or more sequences of DNA when they have overlapping end sequences at their joining point (~40bp). These overlapping regions can be easily added to the ends of any length of DNA by using PCR with primers which have added adapter sequences. Thus PCR followed by Gibson Assembly allows you to join any two blunt ended pieces of DNA. Up to more than 10 different pieces of DNA can be neatly spliced together in a single reaction using these techniques. The technique provides an advance on standard BioBrick assembly, eliminating DNA scars, need for unique restriction sites and providing increased speed.
Gibson Assembly is a high-efficiency DNA end-linking technique developed by Daniel Gibson at the JCVI. The technique was invented and perfected as part of the genome assembly efforts at JCVI. The method uses three enzymes to join two or more sequences of DNA when they have overlapping end sequences at their joining point (~40bp). These overlapping regions can be easily added to the ends of any length of DNA by using PCR with primers which have added adapter sequences. Thus PCR followed by Gibson Assembly allows you to join any two blunt ended pieces of DNA. Up to more than 10 different pieces of DNA can be neatly spliced together in a single reaction using these techniques. The technique provides an advance on standard BioBrick assembly, eliminating DNA scars, need for unique restriction sites and providing increased speed.
In our hands, the method has proved robust and efficient, and has effectively replaced restriction enzyme - DNA ligase style cloning, including standard BioBrick assembly. We have provided an open guide to the technique. We think that the method is of potential use to anyone engaged in DNA assembly and cloning. This is an ongoing effort - we are keen to explore ways of standardising DNA assembly and making biological engineering more efficient. In particular, we are keen to explore low cost education solutions for robotics, reactants and instrumentation. Click here for more details.

The Gibthon Construct Designer is a tool which allows you to automatically generate primers for Gibson Assembly. The Gibthon tool was developed by Bill Collins and the 2010 Cambridge iGEM team to aid, and aprtly automate the design of the oligos for rapid assembly of DNAs .
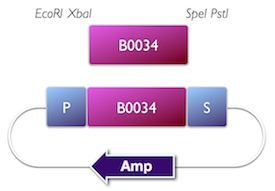
Tom Knight at MIT has pioneered the development of standard DNA parts. He conceived the BioBrick (BBa) format which is the basis for the iGEM competition, and the development of the Registry for Standard Parts. Standardisation has promoted automated, modular approaches to DNA construction, and circuit analysis.
Bioluminescence.
The iGEM2010 team synthesised and refactored a number of bioluminescent components, optimising systems for high level light output. (Ben Reeve and Theo Sanderson are show illuminated by flasks of their engineered luminescent bacteria). The systems show promise for high level light output in plants.
The iGEM2010 team synthesised and refactored a number of bioluminescent components, optimising systems for high level light output. (Ben Reeve and Theo Sanderson are show illuminated by flasks of their engineered luminescent bacteria). The systems show promise for high level light output in plants.

In vivo pigment production as a biomarker.
The biosynthetic pathway for violacein, an intense purple pigment has been engineered for regulated expression in E. coli, based on the work of the Cambridge iGEM2009 team. This is being refactored for expression in plant cells, with the prospect of painting different cell types during plant growth and developemnt.
The biosynthetic pathway for violacein, an intense purple pigment has been engineered for regulated expression in E. coli, based on the work of the Cambridge iGEM2009 team. This is being refactored for expression in plant cells, with the prospect of painting different cell types during plant growth and developemnt.
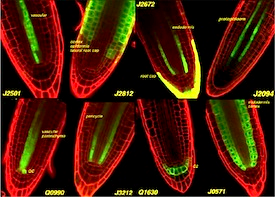
Systems for targeted gene expression in plants.
We have produced a library of transgenic Arabidopsis lines which stably express artificial transcription regulators, based on the GAL4 and HAP1 proteins from yeast. The expression of these regulators is targeted to particular cell types - and can be used to engineer gene expression in those cells. These enhancer trap lines show coordinate expression of a linked GFP or CFP gene. An online catalogue of images is provided. The GAL4 lines are available from the Arabidopsis Stock Centres.
We have produced a library of transgenic Arabidopsis lines which stably express artificial transcription regulators, based on the GAL4 and HAP1 proteins from yeast. The expression of these regulators is targeted to particular cell types - and can be used to engineer gene expression in those cells. These enhancer trap lines show coordinate expression of a linked GFP or CFP gene. An online catalogue of images is provided. The GAL4 lines are available from the Arabidopsis Stock Centres.