Research projects in the Haseloff Laboratory
Synthetic Biology is an emerging field that employs engineering principles to construct new genetic systems. The approach is based on the use of well characterised and reusable DNA components, and numerical models for the design of biological circuits. The approach shows great potential for the engineering of multicellular systems, and plants are the obvious first target for this type of approach. Plants possess indeterminate and modular body plans, have a wide spectrum of biosynthetic activities, can be genetically manipulated, and are used globally for low cost bioproduction at up to gigatonne scale. We are using the lower plant Marchantia polymorpha as a testbed for engineering cellular growth and metabolism. Marchantia is a descendant of the earliest terrestrial plants, and characterised by morphological simplicity, a highly streamlined genome, and a growing set of techniques for facile genetic manipulation, culture and microscopy. Marchantia is providing a new platform for Synthetic Biology and the reprogramming of plant development and physiology.
Work in the Haseloff lab (http://haseloff.plantsci.cam.ac.uk) focuses on the interplay between genetics and plant cell growth, and the way that these interactions can be engineered to give rise to self-organisation and emergent behaviour. We are developing new synthetic biology tools for visualising, manipulating and modelling genetic interactions and morphogenesis. A number of research topics are available for BSc and MEng undergraduate projects, PhD rotation projects or a full programme of PhD work:
Work in the Haseloff lab (http://haseloff.plantsci.cam.ac.uk) focuses on the interplay between genetics and plant cell growth, and the way that these interactions can be engineered to give rise to self-organisation and emergent behaviour. We are developing new synthetic biology tools for visualising, manipulating and modelling genetic interactions and morphogenesis. A number of research topics are available for BSc and MEng undergraduate projects, PhD rotation projects or a full programme of PhD work:

Prototyping genetic circuits in vitro
Recent technical advances in the preparation of microbial cell-free extracts have given rise to a new class of highly efficient systems for gene expression that are cheap to deploy and have huge potential benefit for the provision of a wide variety of diagnostics, sensors, vaccines and research materials. Further, the extracts can be stored desiccated, stable for over a year, and reactivated at the point of use by hydration. The cell free extracts can be programmed by the addition of DNA to allow rapid and simple prototyping of gene circuits for synthetic metabolism, logic and patterning - using production of visible proteins or enzyme products to quantify circuit performance.
A simple plant testbed and modular DNA parts
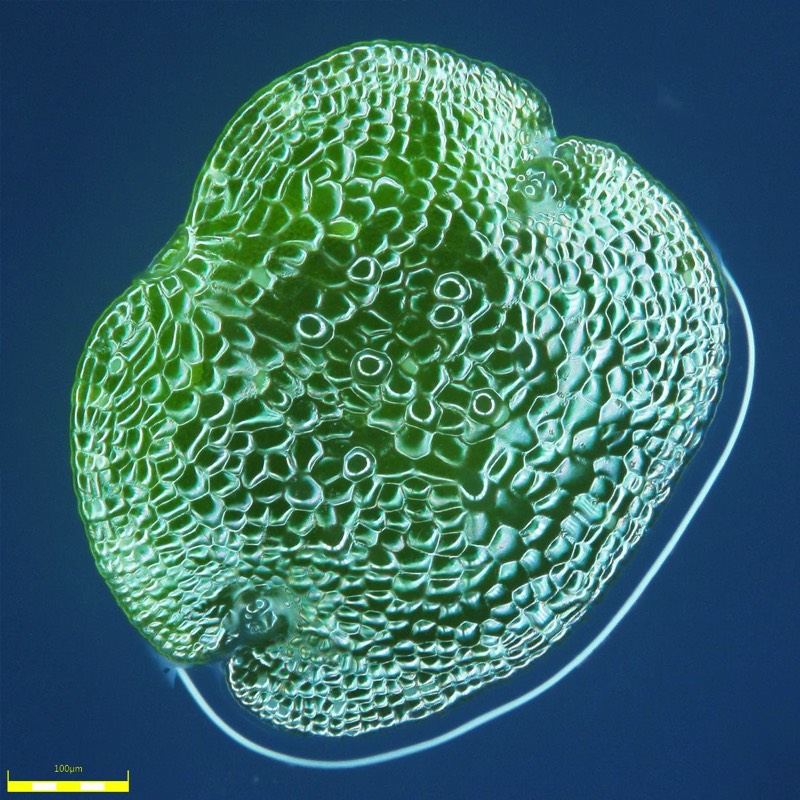
Marchantia polymorpha is a liverwort, descendant of the first terrestrial plants that evolved 500 million years ago. It has a highly simplified body plan, a streamlined genome with all of the genetic mechanisms found in higher plants, and is easy to culture and transform. The plant develops from single-cell spores and undergoes morphogenesis under the microscope. Marchantia is a new testbed for reprogramming of plant development and physiology. As part of the part of the OpenPlant initiative (www.openplant.org), we have established a common syntax for plant DNA parts (Phytobricks), are characterising collections of IP-free DNA parts, and developing techniques for genome-scale DNA editing in this simple plant system. Technical projects are available to characterise DNA parts, including the development of high resolution quantitative microscopy and image processing methods.
Developmental markers and mutants in Marchantia.
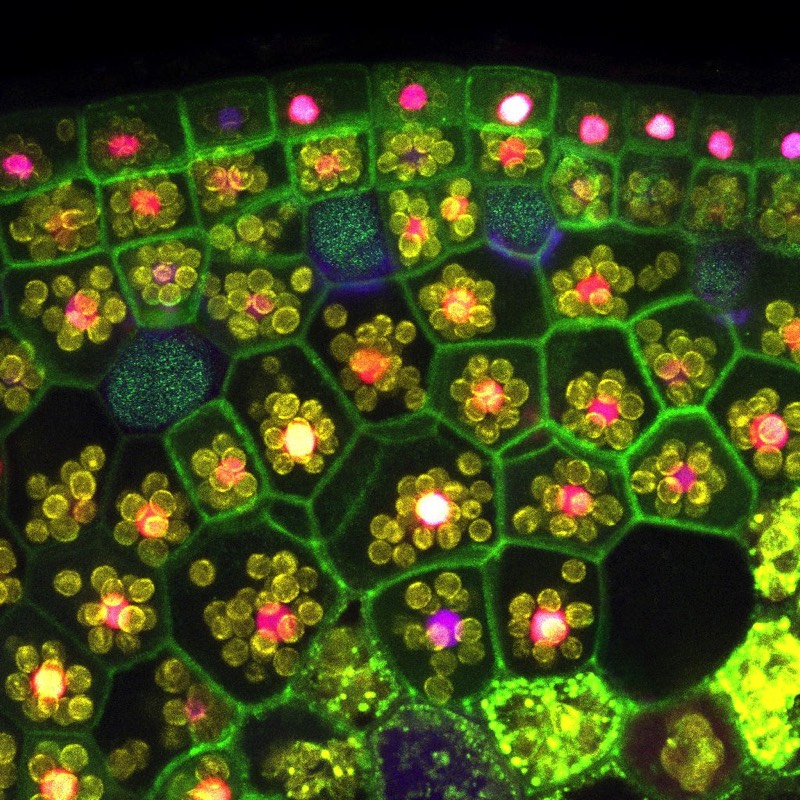
We have established Agrobacterium T-DNA based enhancer trap screens, based on orthogonal transcription factors. In addition we are constructing a library of core promoters from the entire collection of Marchantia transcription factors. These can be used to generate new plant lines which are precisely marked with multispectral fluorescent protein markers. In addition, CRISPR/Cas9 tools can be used to rapidly generate specific gene knock outs. The time course of plant cell proliferation, differentiation and organogenesis can be followed by 3D confocal microscopy. In particular, we are following the development of specialised oil cells and air chambers. The simple genetic architecture and experimental accessibility of Marchantia allows facile study of regulatory systems in situ.
Hormonal control of cell proliferation and plant morphogenesis
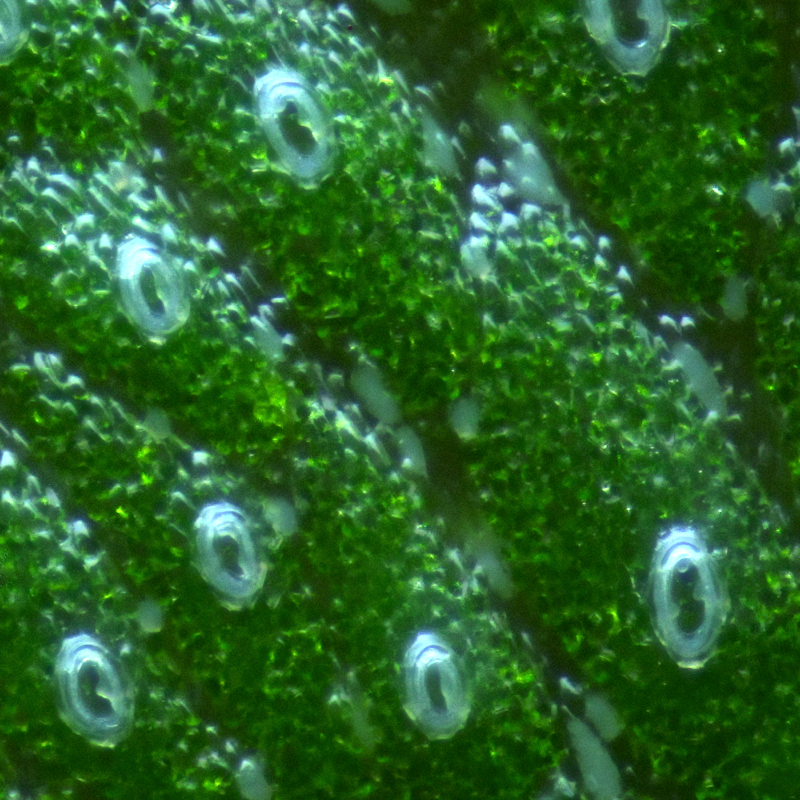
Plant meristems maintain a balance between cells that proliferate and those that are allowed to differentiate. This balance is crucial for proper morphogenesis. Auxins, cytokinins and gibberellins regulate cell division and elongation in plant tissues. Hormonal regulation systems are highly simplified in Marchantia, compared to higher plants. A range of projects are available where targeted expression of genes that regulate auxin, cytokinin and gibberellin levels can be used to alter hormones in situ. The aim of these experiments is to develop generic strategies for engineering growth in Marchantia and other plants.
Chloroplast genome engineering
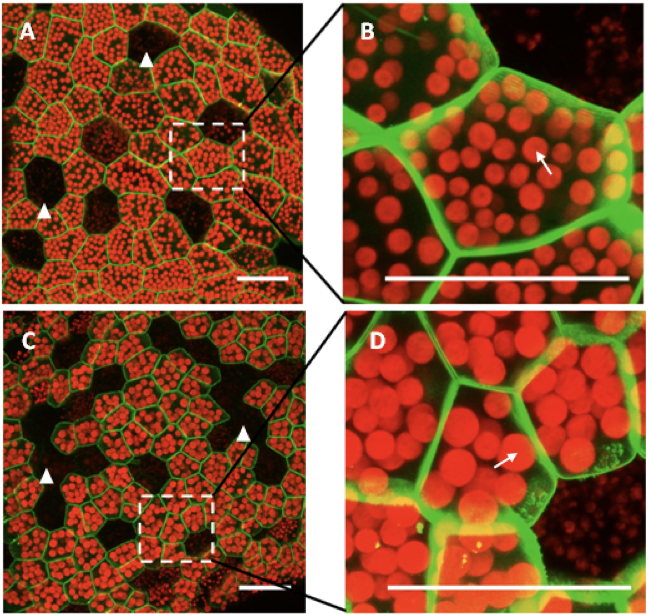
Chloroplasts are major sites for metabolic flux and biosynthesis in plants. There may be 10-100 chloroplasts in each cell, and each plastid contains many copies (10-100) of the relatively small chloroplast genome (121 Kb in Marchantia). Chloroplast transformation has been used for the introduction of various genes and can result in hyperproduction, with the relevant enzyme or antigen sometimes accumulating to a level of more than 50% of total soluble protein. Projects are available for work on the construction of a synthetic chloroplast genome, and establishment of genome transfer and chloroplast engineering techniques in Marchantia.
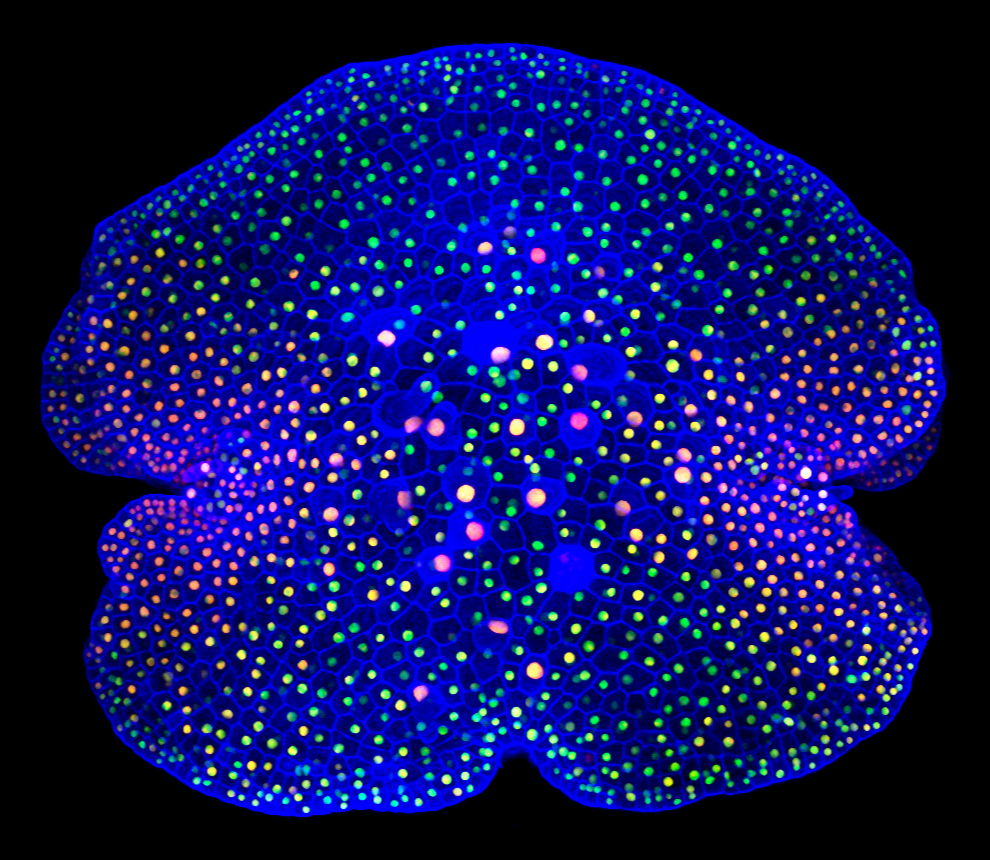
Quantitative 4D imaging of Marchantia development
The prostrate and open form of Marchantia development allows novel approaches to non-invasive, quantitative imaging of the cellular processes that underpin growth. We are developing new microscopy methods, ratiometric markers and automated segmentation techniques for following gene expression, cell division and differentiation in situ. There are opportunities to participate in the development of these new techniques, and to apply them to a number of important scientific questions and engineering challenges.
Tailoring individual projects
Undergraduate and PhD projects in these areas can be interdisciplinary, and tailored to suit a wide range of interests and skills. The listed project areas provide opportunities to work with software models, synthetic DNA circuits and Marchantia (as a simple test bed for engineering multicellular systems). The laboratory is equipped with state-of-the-art robotics, plant culture and imaging equipment. Students will have direct access to modern scientific tools, including confocal microscopes, low light fluorescent imaging and molecular biology equipment, plant and algal growth facilities and computational resources for modelling multicellular growth. The laboratory is a key participant in the OpenPlant initiative (www.openplant.org), and students are encouraged to participate in this and other synthetic biology initiatives in Cambridge (www.synbio.cam.ac.uk). Background information, including downloadable articles and technical information, is available on this web site (http://haseloff.plantsci.cam.ac.uk), and more specific questions can be sent directly to Jim Haseloff (jh295@cam.ac.uk).