iGEM2006 project: Self-organisation
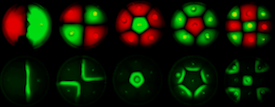
Multicellular organisms undergo self-organisation during development. Our aim was to engineer self-organised pattern formation in free swimming bacteria cells by providing them with an artificial system allowing bi-directional communication. Escherichia coli cells would be equipped with genes derived from independent quorum sensing systems from Pseudomonas aeruginosa and Vibrio fischeri. These systems can enable communication between cell populations and regulated switching between competing cell fates. The negotiation of cell fates within bacterial populations can be visualised precisely by expression of different fluorescent proteins. We further sought to construct biological parts that would render possible the construction of a population based bi-stable switch and modelled the behaviour of single cells as well as the interactive behaviour of populations of cells containing these genetic circuits. Our experiments verified that differential cell motility in combination with position-dependent gene expression has the potential to generate complex patterns.
The Cambridge iGEM2006 wiki is at:
http://2006.igem.org/Team:Cambridge
Background information is at:
http://synbio.org.uk/cambridge/cambridge-igem-teams.html
Multicellular organisms undergo self-organisation during development. Our aim was to engineer self-organised pattern formation in free swimming bacteria cells by providing them with an artificial system allowing bi-directional communication. Escherichia coli cells would be equipped with genes derived from independent quorum sensing systems from Pseudomonas aeruginosa and Vibrio fischeri. These systems can enable communication between cell populations and regulated switching between competing cell fates. The negotiation of cell fates within bacterial populations can be visualised precisely by expression of different fluorescent proteins. We further sought to construct biological parts that would render possible the construction of a population based bi-stable switch and modelled the behaviour of single cells as well as the interactive behaviour of populations of cells containing these genetic circuits. Our experiments verified that differential cell motility in combination with position-dependent gene expression has the potential to generate complex patterns.
The Cambridge iGEM2006 wiki is at:
http://2006.igem.org/Team:Cambridge
Background information is at:
http://synbio.org.uk/cambridge/cambridge-igem-teams.html

iGEM 2006 Students
Kaj Bernhardt: (Biology, Cambridge) & PhD (Germany)
Elizabeth Carter: (Biology, Cambridge) & MBA
Nikhilesh Chand: (Biology, Cambridge) & PhD (Harvard)
Jisun Lee: (Engineering, Cambridge) & PhD Northwestern University (USA)
Yang Xu: (Engineering, Cambridge) & Qualified Engineer
Xueni Zhu: (Engineering, Cambridge) & Qualified Engineer
Kaj Bernhardt: (Biology, Cambridge) & PhD (Germany)
Elizabeth Carter: (Biology, Cambridge) & MBA
Nikhilesh Chand: (Biology, Cambridge) & PhD (Harvard)
Jisun Lee: (Engineering, Cambridge) & PhD Northwestern University (USA)
Yang Xu: (Engineering, Cambridge) & Qualified Engineer
Xueni Zhu: (Engineering, Cambridge) & Qualified Engineer