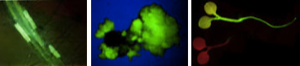
The ß-glucuronidase (GUS) gene is used extensively as a reporter for gene expression in plants. Transformed tissues or patterns of gene expression can be identified histochemically, but this is generally a destructive test and is not suitable for assaying primary transformants, nor for following the time course of gene expression in living plants, nor as a means of rapidly screening segregating populations of seedlings. The green fluorescent protein (GFP) from the cnidarian jellyfish Aequorea victoria shares none of these problems, and we have experimented with its use as a marker in transgenic Arabidopsis thaliana. However, both the expression of GFP within plant cells, and the efficient regeneration of brightly fluorescent transgenic plants has proved problematic.
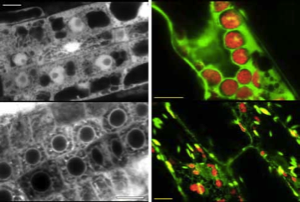
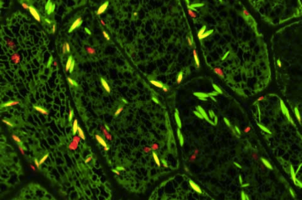
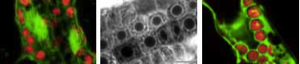
We have shown that proper expression of GFP in the model plant Arabidopsis thaliana is curtailed due to aberrant mRNA processing. An 84 nucleotide cryptic intron is efficiently recognised and removed from within transcripts of the GFP coding sequence, and prevents expression of the protein. We have modified the codon usage of the gfp gene to alter the intron, and restore proper expression in Arabidopsis. GFP was localised within the nucleoplasm and cytoplasm of transformed Arabidopsis cells and could give rise to high levels of fluorescence, but it proved difficult to efficiently regenerate transgenic plants from the brightest cells. The protein may be mildly toxic at high levels, perhaps due to fluorescence-related free radical production. In jellyfish photocytes, GFP is sequestered within cytoplasmic organelles. When GFP was targeted to the endoplasmic reticulum and excluded from the nucleus in Arabidopsis, transformed cells regenerated routinely to give highly fluorescent plants. (Jim Haseloff, Kirby R. Siemering, Douglas Prasher & Sarah Hodge, Removal of a cryptic intron and subcellular localisation of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA, 1997).
Expression of GFP in cells that require higher incubation temperatures has also been problematic. We have also carried out a screen for mutant forms of GFP that fluoresce more intensely when expressed in E. coli at 37°C. We have characterised a bright mutant with reduced sensitivity to temperature in both bacteria and yeast and shown that the amino acid substitutions present act by preventing temperature-dependent misfolding of the apoprotein. We have shown that the fluorescence spectra of the mutant GFP can be manipulated by site-directed mutagenesis without disturbing its improved folding characteristics and have produced a thermostable folding mutant (mgfp5) that can be efficiently excited using either long wavelength UV or blue light. Expression of mgfp5 results in greatly improved levels of fluorescence in both microbial and mammalian cells cultured at 37°C. (Kirby Siemering, Ralph Golbik, Richard Sever & Jim Haseloff, Mutations that supress the thermosensitivity of green fluorescent protein. Current Biology 6:1653-1663, 1996).
The thermotolerant mutants of GFP greatly improve the sensitivity of the protein as a visible reporter molecule in various cell types. The fluorescence spectra of these mutants can be manipulated by further mutagenesis without deleteriously affecting their improved folding characteristics, which introduces the possibility of engineering a range of spectral variants with improved tolerance to temperature. Such a range of sensitive reporter proteins will greatly improve the prospects for GFP-based applications in cells that require higher incubation temperatures.
The modified gfp gene is useful for directly monitoring gene expression and as a simply scored transformation marker in living plants. GFP-expressing seedlings can be examined by confocal microscopy to obtain high resolution optical sections of living tissues, and for time-lapse observation of dynamic processes within cells.
Expression of GFP in cells that require higher incubation temperatures has also been problematic. We have also carried out a screen for mutant forms of GFP that fluoresce more intensely when expressed in E. coli at 37°C. We have characterised a bright mutant with reduced sensitivity to temperature in both bacteria and yeast and shown that the amino acid substitutions present act by preventing temperature-dependent misfolding of the apoprotein. We have shown that the fluorescence spectra of the mutant GFP can be manipulated by site-directed mutagenesis without disturbing its improved folding characteristics and have produced a thermostable folding mutant (mgfp5) that can be efficiently excited using either long wavelength UV or blue light. Expression of mgfp5 results in greatly improved levels of fluorescence in both microbial and mammalian cells cultured at 37°C. (Kirby Siemering, Ralph Golbik, Richard Sever & Jim Haseloff, Mutations that supress the thermosensitivity of green fluorescent protein. Current Biology 6:1653-1663, 1996).
The thermotolerant mutants of GFP greatly improve the sensitivity of the protein as a visible reporter molecule in various cell types. The fluorescence spectra of these mutants can be manipulated by further mutagenesis without deleteriously affecting their improved folding characteristics, which introduces the possibility of engineering a range of spectral variants with improved tolerance to temperature. Such a range of sensitive reporter proteins will greatly improve the prospects for GFP-based applications in cells that require higher incubation temperatures.
The modified gfp gene is useful for directly monitoring gene expression and as a simply scored transformation marker in living plants. GFP-expressing seedlings can be examined by confocal microscopy to obtain high resolution optical sections of living tissues, and for time-lapse observation of dynamic processes within cells.