CellModeller version 4
CellModeller is a framework for modelling multicellular systems, including biophysics, regulatory dynamics, intercellular signalling, and rule-based behaviours. It is implemented using OpenCL, a parallel computation standard, allowing very large numbers of cells to be simulated
Morphogenesis is the process of creating and maintaining form through coordinated cell-division and growth. We have developed a software environment for modelling plant morphogenesis - an interactive virtual laboratory with a front-end called CellModeller4.
We have implemented cellular models of rod-shaped bacteria (3D) and plant tissues (2D). The bacterial model is a novel growing rigid-body method, described here. The plant model is a finite element method based on the work of Lionel Dupuy in the lab, and implemented using a matrix-free approach. The plant model is in the final testing stage, and will be released soon. The bacterial model is a constraint solver, and so can incorporate growth conditions like a flat substrate or microfluidic chamber by including additional constraints.
CellModeller is a framework for modelling multicellular systems, including biophysics, regulatory dynamics, intercellular signalling, and rule-based behaviours. It is implemented using OpenCL, a parallel computation standard, allowing very large numbers of cells to be simulated
Morphogenesis is the process of creating and maintaining form through coordinated cell-division and growth. We have developed a software environment for modelling plant morphogenesis - an interactive virtual laboratory with a front-end called CellModeller4.
We have implemented cellular models of rod-shaped bacteria (3D) and plant tissues (2D). The bacterial model is a novel growing rigid-body method, described here. The plant model is a finite element method based on the work of Lionel Dupuy in the lab, and implemented using a matrix-free approach. The plant model is in the final testing stage, and will be released soon. The bacterial model is a constraint solver, and so can incorporate growth conditions like a flat substrate or microfluidic chamber by including additional constraints.
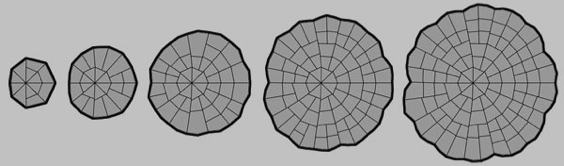
The 2D model is a matrix of linear walls controlled by the adjacent cells. Using the model we can design patterns, shapes and structures by constructing genetic scripts, effectively a kind of genetic sculpture. This is of great value in testing hypotheses about how genes and signalling molecules direct the growth and development of plants. If biological equivalents can be found these could even then be grown by engineering real cellular systems.
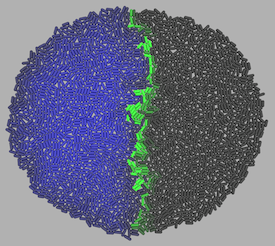
The program has proved robust and the models show lifelike behaviour. We are beginning to test the program, side-by-side with genetic experiments using foreign transcription activators and different coloured fluorescent proteins, to experiment with artificial pattern formation in microbial populations and plant tissues.

Computational modeling of synthetic microbial biofilms Rudge TJ*, Steiner PJ*, Phillips A, and Haseloff J. ACS Synthetic Biology (2012)
Coordination of plant cell division and expansion in a simple morphogenetic system Dupuy, L., Mackenzie, J. and Haseloff, J. Proc. Natl. Acad. Sci. USA 107:2711-6 (2010)
A system for modelling cell-cell interactions during plant morphogenesis Dupuy L, Mackenzie J, Rudge T, Haseloff J. Annals of Botany 101:1255-1265 (2008)
A biomechanical model for the study of plant morphogenesis: Coleochaete orbicularis Dupuy, L.X., Mackenzie, J and Haseloff, J. Proceedings of the 5th Plant Biomechanics Conference. Stockholm. (2006)
A computational model of cellular morphogenesis in plants Rudge, T. and Haseloff, J. Lecture Notes in Computer Science: Advances in Artificial Life, 3630:78-87 (2005)
