Computational analysis and design of morphogenetic circuits in plants.

We are attempting to provide a framework for this. Our aim is to produce: (i) a simplified numerical description of cell arrangements within a meristem, (ii) a related set of algorithms for modelling cell interactions, and (iii) a means of visualising dynamic models for cell development. A numerical description of the plant embryo and meristem will pave the way for the use of biological data for building and testing dynamic models of cellular development.
CellModeller software
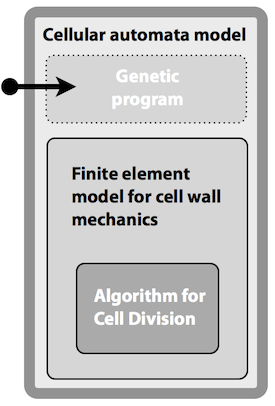
We have developed software tools for dynamically modelling cellular morphogenesis in plants and microbes. The physical properties of cells have been modelled in 2D and 3D, using biophysical models to describe wall properties and cell interactions. The physical model provides an engine for the production of cells through enlargement and division. The CellModeller program provides an environment for combining physical and genetic descriptions of multicellular growth. Fields of proliferating cells can be programmed via a genetic script to produce and respond to different morphogens.
We have developed software tools for dynamically modelling cellular morphogenesis in plants and microbes. The physical properties of cells have been modelled in 2D and 3D, using biophysical models to describe wall properties and cell interactions. The physical model provides an engine for the production of cells through enlargement and division. The CellModeller program provides an environment for combining physical and genetic descriptions of multicellular growth. Fields of proliferating cells can be programmed via a genetic script to produce and respond to different morphogens.
Design of self-organising genetic circuits
We are using microbial and plant systems to test designs for genetic circuits that, once initiated, produce stable spatio-temporal patterns of gene expression in cell assemblages. The circuits are based the ideas of Alan Turing, where diifferential traffic of signalling molecules and feedback can be harnessed to create ordered systems.
We are using microbial and plant systems to test designs for genetic circuits that, once initiated, produce stable spatio-temporal patterns of gene expression in cell assemblages. The circuits are based the ideas of Alan Turing, where diifferential traffic of signalling molecules and feedback can be harnessed to create ordered systems.
Biophysical models for cellular growth in plants
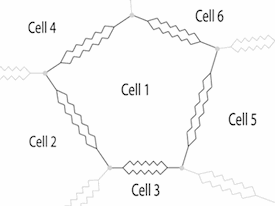
The dynamics of cellular morphogenesis in plants can be modelled using software. The physical properties of cells have been modelled in 2D, using viscosity to describe wall properties. This physical model provides an engine for the production of cells through enlargement and division.
The dynamics of cellular morphogenesis in plants can be modelled using software. The physical properties of cells have been modelled in 2D, using viscosity to describe wall properties. This physical model provides an engine for the production of cells through enlargement and division.
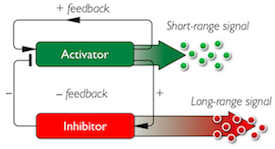
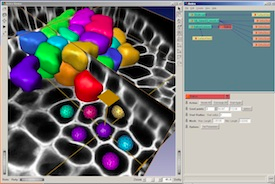
Image processing
Classical botanical microtechniques can be adapted for modern laser scanning confocal microscopy, to allow deep optical sectioning of intact plant tissues. The resulting large datasets can be visualised and converted to simple numerical descriptions of cell arrangement, shape and connectivity using 3D segmentation software. A similar approach can be taken for living tissues, using genetic probes, and image processing techniques provide a window into the dynamic properties of plant and microbial tissues.
Classical botanical microtechniques can be adapted for modern laser scanning confocal microscopy, to allow deep optical sectioning of intact plant tissues. The resulting large datasets can be visualised and converted to simple numerical descriptions of cell arrangement, shape and connectivity using 3D segmentation software. A similar approach can be taken for living tissues, using genetic probes, and image processing techniques provide a window into the dynamic properties of plant and microbial tissues.