A Quick Guide to Oligonucleotide Design for PCR and Gibson End-linked DNA Assembly
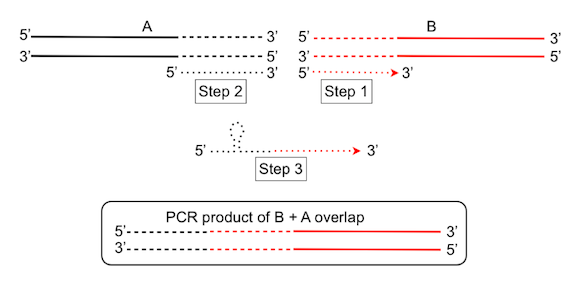
The design of an oligo primer pair for PCR and Gibson based end-linked DNA assembly can be summarized in three stages. In this example the process will join two fragments, denoted A and B in the figure below.
Since sequences from the database are conventionally in the 5’ to 3’ direction and DNA polymerases only work 5’ to 3’, for this example the sequence can be taken directly from a Genbank file for the primer design. In total, about 40 bp of overlap should be sufficient for the Gibson assembly reaction after PCR.
Stage 1. From the 5’ end of fragment B sequence, go in the 3’ direction about 20 bp. In the first instance, take 20 bp and then adjust to try and optimize the 3’ anchoring residues (preferably G or C) and check the Tm. Over 20 bp, the Tm is generally fine unless the sequence is heavily GC or AT rich. Remember that the Tm for the other PCR primer should be about the same. A primer design program such as Primer3 (http://frodo.wi.mit.edu/primer3) or visual OMPTM (http://dnasoftware.com), are helpful, if the exact position of the overlap is not constrained, for example, when a long sequence needs to be broken into two or more smaller sequences for efficient PCR.
Stage 2. From the 3’ end of fragment A, sequence, go in the 5’ direction about 20 bp.
Stage 3. Join these two sequences and check for single stranded DNA secondary structure using a program like Unafold or MFold (http://mfold.bioinfo.rpi.edu/). Inevitably there will be some stem-loop structures but what matters is that they are not stable (e.g. negative ΔG) and try to avoid strong structure at the 3’ end as this is where the primer must anneal for the PCR. The primer design may have to go through a few iterations in extending or shortening the sequence at either end to achieve a good theoretical primer. The complementary sequence may be a good starting point for the fragment A primer to generate the overlap with fragment B.
NB: Check the reverse complement of the sequence from Step 3 as it will be a good starting point for the fragment A PCR primer that will generate an overlap with fragment B.