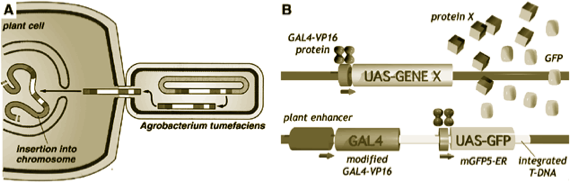
GAL4-GFP enhancer trap system.
A system for GAL4 targeted gene expression. An enhancer trap vector bearing a modified GAL4-VP16 gene was inserted randomly into the Arabidopsis genome by Agrobacterium mediated transformation. Cell specific activation of the GAL4-VP16 gene by cellular enhancer results in activation of a linked GFP gene, allowing simple characterisation of expression patterns. Targeted expression of another gene (X) can be induced by genetic crossing. For example, we can use the system to trigger cell ablation by targeted expression of a toxin.
A system for GAL4 targeted gene expression. An enhancer trap vector bearing a modified GAL4-VP16 gene was inserted randomly into the Arabidopsis genome by Agrobacterium mediated transformation. Cell specific activation of the GAL4-VP16 gene by cellular enhancer results in activation of a linked GFP gene, allowing simple characterisation of expression patterns. Targeted expression of another gene (X) can be induced by genetic crossing. For example, we can use the system to trigger cell ablation by targeted expression of a toxin.
The GAL4 system
In order to genetically manipulate cells during meristem development, we have developed a scheme for targeted gene expression, which is based on a method widely used in Drosophila (Brand and Perrimon, Development 118:401-415, 1993). We have used an "enhancer-trap" strategy to generate many transgenic plants which express different patterns of a yeast transcription activator, GAL4. A chosen target gene can then be placed under the control of GAL4 upstream activation sequences (UAS), transformed into plants, and maintained silently in the absence of GAL4. Genetic crosses between this single line and any of the library of GAL4-containing lines specifically activates the target gene in a particular tissue or cell type. The phenotypic consequences of mis-expression, including those lethal to the organism, can be conveniently studied.We found that GAL4 is not expressed in Arabidopsis due to a high A/T content, which can interfere with mRNA processing in plants. We have altered the codon usage of a derivative, GAL4-VP16, so that it is expressed efficiently in plants, and randomly inserted the modified gene into the Arabidopsis genome, using Agrobacterium tumefaciens-mediated transformation. The transformation vector was designed so that expression of the GAL4-VP16 gene would be dependent upon the fortuitous proximity of an Arabidopsis enhancer element. The inserted DNA also contained a GAL4-responsive green fluorescent protein (GFP) gene. Thus, interesting "enhancer-trap" patterns of GAL4 gene expression were immediately and directly visible, with each GAL4-expressing cell marked by bright green fluorescence.
We have used in vivo detection of GFP to directly screen for GAL4-directed GFP expression in 7500 regenerated plantlets. We have documented a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root. These lines provide a valuable set of markers, where particular cell types are tagged and can be visualised with unprecedented ease and clarity in living plants. Most importantly, GAL4-VP16 expression within these lines can be used for targeted gene expression experiments. We have produced transgenic plants which maintain regulatory proteins or toxins, silent behind a GAL4-responsive promoter. We can now activate these genes in specific cells by crossing to a chosen GAL4-VP16 expressing line.
In order to genetically manipulate cells during meristem development, we have developed a scheme for targeted gene expression, which is based on a method widely used in Drosophila (Brand and Perrimon, Development 118:401-415, 1993). We have used an "enhancer-trap" strategy to generate many transgenic plants which express different patterns of a yeast transcription activator, GAL4. A chosen target gene can then be placed under the control of GAL4 upstream activation sequences (UAS), transformed into plants, and maintained silently in the absence of GAL4. Genetic crosses between this single line and any of the library of GAL4-containing lines specifically activates the target gene in a particular tissue or cell type. The phenotypic consequences of mis-expression, including those lethal to the organism, can be conveniently studied.We found that GAL4 is not expressed in Arabidopsis due to a high A/T content, which can interfere with mRNA processing in plants. We have altered the codon usage of a derivative, GAL4-VP16, so that it is expressed efficiently in plants, and randomly inserted the modified gene into the Arabidopsis genome, using Agrobacterium tumefaciens-mediated transformation. The transformation vector was designed so that expression of the GAL4-VP16 gene would be dependent upon the fortuitous proximity of an Arabidopsis enhancer element. The inserted DNA also contained a GAL4-responsive green fluorescent protein (GFP) gene. Thus, interesting "enhancer-trap" patterns of GAL4 gene expression were immediately and directly visible, with each GAL4-expressing cell marked by bright green fluorescence.
We have used in vivo detection of GFP to directly screen for GAL4-directed GFP expression in 7500 regenerated plantlets. We have documented a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root. These lines provide a valuable set of markers, where particular cell types are tagged and can be visualised with unprecedented ease and clarity in living plants. Most importantly, GAL4-VP16 expression within these lines can be used for targeted gene expression experiments. We have produced transgenic plants which maintain regulatory proteins or toxins, silent behind a GAL4-responsive promoter. We can now activate these genes in specific cells by crossing to a chosen GAL4-VP16 expressing line.
Modified GAL4-VP16 sequence
The coding sequence for the GAL4 DNA binding domain is A:T rich and is not well expressed in Arabidopsis. We have extensively modified its codon usage, and constructed a modified GAL4-VP16 gene for improved plant expression.
The coding sequence for the GAL4 DNA binding domain is A:T rich and is not well expressed in Arabidopsis. We have extensively modified its codon usage, and constructed a modified GAL4-VP16 gene for improved plant expression.
Enhancer-trap screen
Our particular interest is in the cells of the Arabidopsis root tip, and we have used in vivo detection of GFP to develop a new and efficient GAL4 enhancer-trap screening procedure. At the beginning of this work, it was not feasible for us to handle many thousands of Arabidopsis plant lines in our small greenhouse, and we used in vivo detection of GFP to develop a new and efficient screening procedure. As our particular interest is in the cells of the Arabidopsis root tip, we modified a tissue culture-based plant transformation protocol to include an auxin induction of roots from regenerating shootlets. More than 7500 transformants were then generated, planted in grid patterns in sterile culture dishes and directly screened for GAL4/GFP expression within roots. Several hundred lines with interesting patterns of root expression were choosen, documented, transferred to soil and grown to seed, to both amplify and self-hybridise the lines. As a result, we produced a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root. These were made available through the Arabidopsis Stock Centre, and we have created a graphical database of the expression patterns to allow easy computer and web access at this site. These lines provide a valuable set of markers, where particular cell types are tagged and can be visualised with unprecedented ease and clarity in living plants.
Our particular interest is in the cells of the Arabidopsis root tip, and we have used in vivo detection of GFP to develop a new and efficient GAL4 enhancer-trap screening procedure. At the beginning of this work, it was not feasible for us to handle many thousands of Arabidopsis plant lines in our small greenhouse, and we used in vivo detection of GFP to develop a new and efficient screening procedure. As our particular interest is in the cells of the Arabidopsis root tip, we modified a tissue culture-based plant transformation protocol to include an auxin induction of roots from regenerating shootlets. More than 7500 transformants were then generated, planted in grid patterns in sterile culture dishes and directly screened for GAL4/GFP expression within roots. Several hundred lines with interesting patterns of root expression were choosen, documented, transferred to soil and grown to seed, to both amplify and self-hybridise the lines. As a result, we produced a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root. These were made available through the Arabidopsis Stock Centre, and we have created a graphical database of the expression patterns to allow easy computer and web access at this site. These lines provide a valuable set of markers, where particular cell types are tagged and can be visualised with unprecedented ease and clarity in living plants.
Enhancer trap scheme with GFP
1. A gene for a foreign transcription activator, mGAL4-VP16 gene (blue), is inserted into the Arabidopsis genome (red) via Agrobacterium mediated transformation.
2. The proximity of cellular enhancer elements can drive expression of mGAL4-VP16 in distinct temporal and spatial patterns in the plant.
3. The GAL4-VP16 protein will activate transcription from a promoter containing GAL4 upstream activator sequences (UAS). A linked GAL4-responsive mgfp5-ER gene (green) marks the GAL4-expressing cells with green fluorescence. This enables primary transformants to be directly screened for patterns of GAL4 expression.
4. Any chosen gene(s) can then be crossed with a GAL4-GFP expressing line, and specifically transactivated in the GAL4-expressing cells.
1. A gene for a foreign transcription activator, mGAL4-VP16 gene (blue), is inserted into the Arabidopsis genome (red) via Agrobacterium mediated transformation.
2. The proximity of cellular enhancer elements can drive expression of mGAL4-VP16 in distinct temporal and spatial patterns in the plant.
3. The GAL4-VP16 protein will activate transcription from a promoter containing GAL4 upstream activator sequences (UAS). A linked GAL4-responsive mgfp5-ER gene (green) marks the GAL4-expressing cells with green fluorescence. This enables primary transformants to be directly screened for patterns of GAL4 expression.
4. Any chosen gene(s) can then be crossed with a GAL4-GFP expressing line, and specifically transactivated in the GAL4-expressing cells.

Agrobacterium-mediated transformation
Agrobacterium-mediated transformation is used to transform plant cells with a plasmid containing the modified GAL4-VP16 and UAS-GFP genes. The modified GAL4-VP16 gene is positioned adjacent to the Tr border sequence with the truncated (-48) 35S promoter. Transformation results in integration of the T-DNA at random positions into the plant genome. Proper expression of the GAL4-VP16 gene in these transformed plants requires the fortuitous presence of plant enhancer elements in flanking chromosomal sequences. Expression of the GAL4-VP16 gene then drives the GFP reporter gene.
Agrobacterium-mediated transformation is used to transform plant cells with a plasmid containing the modified GAL4-VP16 and UAS-GFP genes. The modified GAL4-VP16 gene is positioned adjacent to the Tr border sequence with the truncated (-48) 35S promoter. Transformation results in integration of the T-DNA at random positions into the plant genome. Proper expression of the GAL4-VP16 gene in these transformed plants requires the fortuitous presence of plant enhancer elements in flanking chromosomal sequences. Expression of the GAL4-VP16 gene then drives the GFP reporter gene.
Plant transformation
Roots of Arabidopsis thaliana ecotype C24 were transformed using essentially the protocol of Valvekens, D., Van Montagu, M. & Van Lijsebettens, M. (1988) Proc. Natl. Acad. Sci. USA 85, 5536-5540. Root material was first harvested from seedlings grown in sterile liquid culture. Root pieces were treated with 2,4D to induce callus formation. These were then co-cultivated with Agrobacterium tumefaciens strain LBA4044 containing the GAL4-GFP enhancer trap plasmid. The treated material was then transferred to agar media containing selective antibiotics and isopentyl adenine (IPA) for induction of shoot formation. After green shoots had formed on the calli (4-6 weeks), shoots were transferred to media containing isobutyric acid (IBA) for induction of roots.
Roots of Arabidopsis thaliana ecotype C24 were transformed using essentially the protocol of Valvekens, D., Van Montagu, M. & Van Lijsebettens, M. (1988) Proc. Natl. Acad. Sci. USA 85, 5536-5540. Root material was first harvested from seedlings grown in sterile liquid culture. Root pieces were treated with 2,4D to induce callus formation. These were then co-cultivated with Agrobacterium tumefaciens strain LBA4044 containing the GAL4-GFP enhancer trap plasmid. The treated material was then transferred to agar media containing selective antibiotics and isopentyl adenine (IPA) for induction of shoot formation. After green shoots had formed on the calli (4-6 weeks), shoots were transferred to media containing isobutyric acid (IBA) for induction of roots.
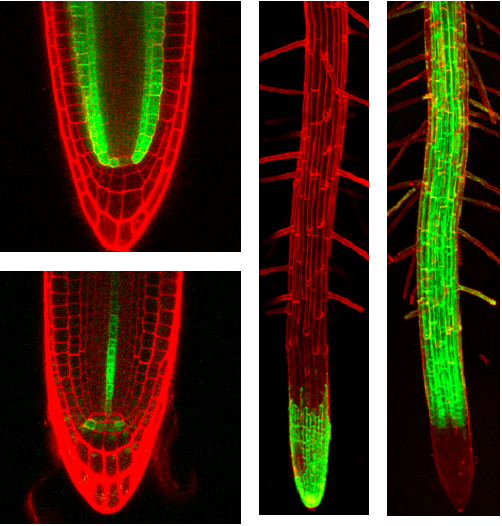
Microscopy
Transgenic plantlets were then screened for GFP expression using a inverted fluorescence microscope (Leitz DM-IL) fitted with a filter set (Leitz-D excitation BP355-425, dichroic 455, emission LP460 ) suitable for the main 395nm excitation and 509nm emission peaks of GFP. The use of a 7mm threaded extension tube with a 4x objective (EF 4/0.12) gave a greater working distance, and allowed the convenient direct observation of shoots within inverted petri dishes. Expression patterns were documented using a Sony 3-CCD colour video camera and integrating frame store. More than 7500 primary transformants were generated, and several hundred lines with interesting patterns of root expression were directly transferred to magenta pots and grown to seed, to both amplify and self-hybridise the lines. We have a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root.
Transgenic plantlets were then screened for GFP expression using a inverted fluorescence microscope (Leitz DM-IL) fitted with a filter set (Leitz-D excitation BP355-425, dichroic 455, emission LP460 ) suitable for the main 395nm excitation and 509nm emission peaks of GFP. The use of a 7mm threaded extension tube with a 4x objective (EF 4/0.12) gave a greater working distance, and allowed the convenient direct observation of shoots within inverted petri dishes. Expression patterns were documented using a Sony 3-CCD colour video camera and integrating frame store. More than 7500 primary transformants were generated, and several hundred lines with interesting patterns of root expression were directly transferred to magenta pots and grown to seed, to both amplify and self-hybridise the lines. We have a collection of 250 Arabidopsis lines with distinct and stable patterns of GAL4-VP16 and GFP expression in the root.
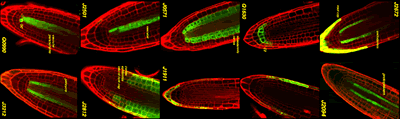
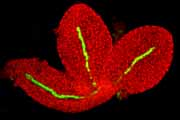
Selected Arabidopsis GAL4-VP16 enhancer trap lines.
Confocal micrographs of selected enhancer trap lines, showing root tip specific expression. The roots were counterstained with propidium iodide (red channel). The GAL4-driven GFP signal is shown superimposed (green channel).
Confocal micrographs of selected enhancer trap lines, showing root tip specific expression. The roots were counterstained with propidium iodide (red channel). The GAL4-driven GFP signal is shown superimposed (green channel).