A computational system for modelling cellular morphogenesis in plants
In the first steps towards developing a system for dynamic modelling of plant cell development, we have generated a 2D description of the physical properties of cells, using a novel double spring model to describe cell wall properties. This physical model provides an engine for the production of cells through enlargement and division. Fields of proliferating cells can then be programmed via a genetic script to produce and respond to different morphogens. We are using this 2D system as a pilot study for modelling cell dynamics and interactions in 3D. However it is also proving very useful as a tool for modelling interactions within single layers of cells, such as the epidermis, and for modelling zonation within a meristem.
In the first steps towards developing a system for dynamic modelling of plant cell development, we have generated a 2D description of the physical properties of cells, using a novel double spring model to describe cell wall properties. This physical model provides an engine for the production of cells through enlargement and division. Fields of proliferating cells can then be programmed via a genetic script to produce and respond to different morphogens. We are using this 2D system as a pilot study for modelling cell dynamics and interactions in 3D. However it is also proving very useful as a tool for modelling interactions within single layers of cells, such as the epidermis, and for modelling zonation within a meristem.
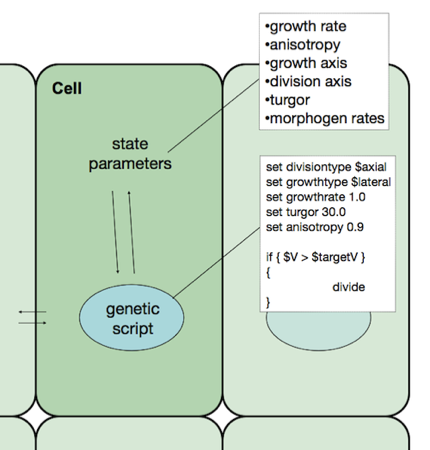
Cell parameters
The parameters which define the state of each cell and which are available for transformation by the genetic script are as follows:
Growth rate: The relative rate of growth of the cell defined on [0,1].
Anisotropy: The degree of polarity in cell growth ranging from zero (isotropic growth) to one (growth restricted to direction of polarity)
Growth axis: The direction along which anisotropic growth will occur most. This is defined relative to the dividing wall which created the cell, and may be parallel or perpendicular to it.
Division axis: The direction along which the dividing wall is formed during mitosis. Again this is defined relative to the dividing wall which created the cell.
Turgor: The internal turgor of a cell is modelled to allow it to maintain its shape and to allow the transmission of hydrostatic pressure across the tissue.
Morphogen concentrations: These form the basis of the chemical patterns which may then influence cell behaviour via the genetic script. This is analogous to gene expression in response to signalling molecules.
Morphogen production rates: The rate at which each cell is producing or degrading each of the morphogens in the system.
Timer: A timer variable is initialised to zero when a cell divides: analogous to the cell cycle.
Volume/Size: In 2D this is the area enclosed by the cell boundary. This may not be directly changed by the script but is affected by altering the growth parameters.The boundary of each cell is defined by a set of linear walls which form a closed polygon. The geometry of the walls is controlled by our spatial model.
The parameters which define the state of each cell and which are available for transformation by the genetic script are as follows:
Growth rate: The relative rate of growth of the cell defined on [0,1].
Anisotropy: The degree of polarity in cell growth ranging from zero (isotropic growth) to one (growth restricted to direction of polarity)
Growth axis: The direction along which anisotropic growth will occur most. This is defined relative to the dividing wall which created the cell, and may be parallel or perpendicular to it.
Division axis: The direction along which the dividing wall is formed during mitosis. Again this is defined relative to the dividing wall which created the cell.
Turgor: The internal turgor of a cell is modelled to allow it to maintain its shape and to allow the transmission of hydrostatic pressure across the tissue.
Morphogen concentrations: These form the basis of the chemical patterns which may then influence cell behaviour via the genetic script. This is analogous to gene expression in response to signalling molecules.
Morphogen production rates: The rate at which each cell is producing or degrading each of the morphogens in the system.
Timer: A timer variable is initialised to zero when a cell divides: analogous to the cell cycle.
Volume/Size: In 2D this is the area enclosed by the cell boundary. This may not be directly changed by the script but is affected by altering the growth parameters.The boundary of each cell is defined by a set of linear walls which form a closed polygon. The geometry of the walls is controlled by our spatial model.