
Arabidopsis thaliana grows quickly, produces prolific seed, and is easy to transform. Its genome is completely sequenced and a large variety of experimental tools and genetic resources are available. The root meristem grows indeterminately, has a simple and transparent 3D architecture, and can be induced to form de novo in adult tissues. We have developed a combination of new genetic and microscopy techniques for Arabidopsis in order to visualise cell interactions and gene expression within plant tissues, and to reprogram plant gene expression.
The Arabidopsis thaliana genome.
Nucleotide sequence determination of the Arabidopsis thaliana genome is complete and the entire complement of around 30,000 encoded genes is available for inspection. Problems in understanding plant development and growth have been recast. Gene description is no longer an issue. Computational analysis of the Arabidopsis genome has led to the identification and classification of entire gene families. Rather than work solely with the small group of genes that have been identified by genetic or biochemical studies to affect a particular biological process, workers can now aim to build a more complete picture of gene interactions using genome-wide analytical tools. In this post-genomic work, the objective is to assign functions to genes, and to gain a better understanding of how networks of interacting proteins can lead to the assembly and maintenance of the organism. In turn, this knowledge might be used to reprogram plant development.
We work with the Arabidopsis root meristem as a model system. We are developing genetic tools for visualising and manipulating cells within the intact plant, and the simple and well characterised architecture of the root (Dolan et al. Development 119:71-84, 1993; Scheres et al., Development 120:2475-2487, 1994) aids analysis of any perturbation brought about by experimental manipulation. The root meristem consists of a plate of quiescent cells surrounded by initials that divide to produce distal root cap cells, and also lay down continuous cell files proximally. Behind the tip, the newly formed cells of the root undergo differentiation and expansion to build a regular arrangement of cell types within the mature root.
GAL4-GFP Arabidopsis lines.
We have regenerated and screened over 8000 T-DNA transformed Arabidopsis plantlets for enhancer-trap driven GAL4 + UAS:mGFP5-ER expression. (see GAL4 and GFP sections for further information). Several hundred of these GAL4-GFP lines show distinct and stable expression, and we have made them have available through the Arabidopsis stock centres over the last few years. Cells within the meristem can be precisely and easily observed using GFP markers and confocal microscopy of living roots. GAL4-mediated misexpression can be used to genetically manipulate particular cells.
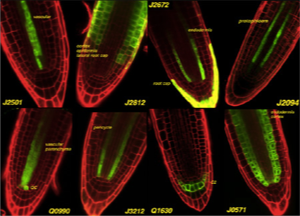
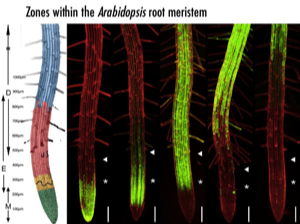
A simple cellular interaction is found the outside of the Arabidopsis root meristem and appears to control a fundamental transition between cell division and elongation. The lateral root cap is laid down by division of initials on the outside of the meristem, and this forms a short layer, a single cell deep, on the outside of the root tip as the oldest of the cells eventually undergo programmed cell death. The epidermis lies immediately underneath this layer. At the tip, epidermal cells undergo a rapid set of 4 or 5 divisions, followed by the rapid onset of cell expansion. We have found that, in Arabidopis, the boundary between dividing and elongating cells in the epidermis is sharp, and always lies adjacent to the terminal root cap cell in the next layer. In addition, we have a number of GAL4 enhancer trap lines that clearly mark this boundary . It has been possible to genetically ablate these cells. When the lateral root cap cells are ablated, epidermal cells undergo accelerated differentiation: cell expansion and the formation of differentiated root hair cells are seen immediately adjacent to the root tip. This indicates that lateral root cap cells play a direct role in maintaining the properties of the rapidly dividing epidermal cells that they contact, and suggests that an interaction between two layers is required for the coordination of cell division and root growth. This simple interaction of cell types is an ideal starting point for model building.
We have used GAL4 targeted gene expression to kill cells and to trigger ectopic cell divisions in the lateral root cap and epidermal layers of the meristem. For example, using an Arabidopis line Q0171 with lateral root cap expression of GAL4, it has been possible to target the diphtheria toxin A chain to these cells, to specifically kill them. When the lateral root cap cells are ablated, epidermal cells are found to undergo accelerated differentiation. This indicates that lateral root cap cells may play a direct role in maintaining the properties of the rapidly dividing epidermal cells that they contact, and suggests that an interaction between two layers is required for the coordination of cell division and root growth.
Cells must adopt particular identities during the construction of a regular body plan in embryogenesis. Every cell is produced by cell division, and adoption of any new fate could be governed by (1) its parent cell (i.e. the cell’s lineage) or (2) the position of the cell within the embryo. The first model would rely on the regular asymmetric segregation of cell fate determinants at cell division during development. While such a mechanism can be seen in some animal systems, proper cell fate determination in Arabidopsis embryos appears independent of regular cell division patterns. Therefore it is more likely that daughter plant cells sense their different positions within the tissue and develop according to regulatory signals exchanged between neighbouring cell.
In Drosophila, genetic screens identified developmental regulators that are required to specify large portions of the animal - and specification of the embryo is increasingly defined during segmentation. In addition, cell fate determinants are found to be asymmetrically segregated within the syncitial embryo and during formative divisions in nervous system development. In contrast, a number of the large scale Arabidopsis patterning defects that have been characterised are due to mutations that affect basic cell processes, such as cytokinesis and cell wall deposition. Disordered cell divisions do not necessarily alter cell fate patterning. It is likely that local cell interactions play a predominant role in determining plant cell fate.
While the exact nature of this positional signalling is unknown, there are several likely mechanisms. (i) Secreted proteins such as CLAVATA3 have been shown to regulate the behaviour of neighbouring cells within a meristem. The target cells possess cell surface receptors that mediate intercellular signalling. (ii) Cell polarity and fate is controlled through the polar transport of the plant hormone auxin between cells, in a feedback dependent process. (iii) Regulatory molecules, including plant transcription factors, have been shown to traffic between cells via plasmodesmata. (iv) Plant cell walls possess specific epitopes which differ between cell types, and which could be used to provide positional cues. Plant meristems possess self-organising properties which appear to be based upon short-range, dynamic exchange of positional information.
Web resources:



TAIR: The Arabidopsis Information Resource
http://www.arabidopsis.org/
The Arabidopsis Information Resource (TAIR) maintains a database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana
http://www.arabidopsis.org/
The Arabidopsis Information Resource (TAIR) maintains a database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana
Lehle Seeds
http://www.arabidopsis.com/
Lehle Seeds, Round Rock, Texas - commercial supplier of Arabidopsis seeds and materials, and sponsor of Arabidopsis-related resources on the arabidopsis.com website.
http://www.arabidopsis.com/
Lehle Seeds, Round Rock, Texas - commercial supplier of Arabidopsis seeds and materials, and sponsor of Arabidopsis-related resources on the arabidopsis.com website.
NASC: Nottingham Arabidopsis Stock Centre
http://arabidopsis.info/
The Nottingham Arabidopsis Stock Centre (NASC) provides seed and information resources to the International Arabidopsis Genome Programme and the wider research community.
http://arabidopsis.info/
The Nottingham Arabidopsis Stock Centre (NASC) provides seed and information resources to the International Arabidopsis Genome Programme and the wider research community.