Computer modelling of morphogenesis in Coleochaete
Extracting Numerical Models from Biological Experiments.
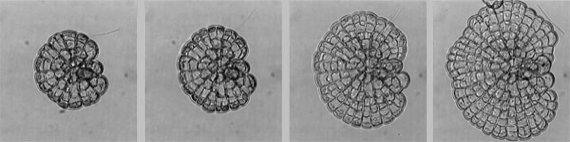
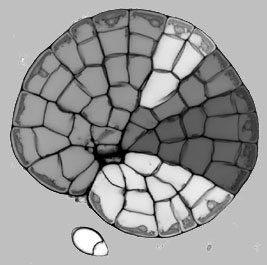
We have developed a number of specific staining procedures for live and fixed Coleochaete colonies, adapted from our work with Arabidopsis. For example, we can precisely label cell walls with a highly fluorescent dye, and capture high contrast images that outline all cells within a colony. We have developed a novel segmentation technique for producing a two dimensional description of the geometry of cells within a Coleochaete colony. (i) A suitable image is randomly seeded with points; (ii) lines are drawn between a point and its neighbours; (iii) pairs of points are classified on the basis of whether the line drawn between them crosses a region of high intensity (a cell wall); (iv) points within the same cell are removed until there is one point remaining per cell; (v) the points are used to initialise a watershed segmentation algorithm; (vi) vertices are positioned at the intersections of multiple watersheds, and these are joined by the cell wall segments. The resulting geometry is saved in an XML-based file structure that we have developed for the storage and exchange of plant cell microarchitecture.
We have developed a number of specific staining procedures for live and fixed Coleochaete colonies, adapted from our work with Arabidopsis. For example, we can precisely label cell walls with a highly fluorescent dye, and capture high contrast images that outline all cells within a colony. We have developed a novel segmentation technique for producing a two dimensional description of the geometry of cells within a Coleochaete colony. (i) A suitable image is randomly seeded with points; (ii) lines are drawn between a point and its neighbours; (iii) pairs of points are classified on the basis of whether the line drawn between them crosses a region of high intensity (a cell wall); (iv) points within the same cell are removed until there is one point remaining per cell; (v) the points are used to initialise a watershed segmentation algorithm; (vi) vertices are positioned at the intersections of multiple watersheds, and these are joined by the cell wall segments. The resulting geometry is saved in an XML-based file structure that we have developed for the storage and exchange of plant cell microarchitecture.

Physical Continuum Model for Tissue Growth.
We have developed a software model to describe the cellular and viscoelastic physical properties of plant tissues. The extracellular matrix of a growing tissue is considered as a continuum, where the activities of individual cells contribute to local differences in turgor and viscoelastic wall properties, and the deformation of the entire growing tissue is solved using finite element methods.
In Silico Morphogenesis of Coleochaete.
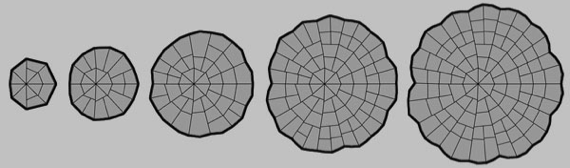
We have combined the viscoelastic model for tissue growth with a cellular automata system and GUI, to produce a software environment for general purpose modelling of plant morphogenetics. Our first task has been to produce working models for the growth of Coleochaete colonies. This requires the assembly of a set of genetic rules that constitute the organism’s genome, and which govern the dynamic behaviour of the colonies. Using a simple set of assumptions, partly based on the empirical rules for cell division formulated in the 1800’s, we have realistically modeled the behaviour of colonies during undisturbed growth, after damage of the meristematic layer, and on the collision of adjacent colonies. In addition, we can successfully extract a description of a Coleochaete colony from a microscope image, and feed this description into the computer modelling software. This provides an important and crucial link between biological experiments and computer modelling.
We have developed a software model to describe the cellular and viscoelastic physical properties of plant tissues. The extracellular matrix of a growing tissue is considered as a continuum, where the activities of individual cells contribute to local differences in turgor and viscoelastic wall properties, and the deformation of the entire growing tissue is solved using finite element methods.
In Silico Morphogenesis of Coleochaete.
We have combined the viscoelastic model for tissue growth with a cellular automata system and GUI, to produce a software environment for general purpose modelling of plant morphogenetics. Our first task has been to produce working models for the growth of Coleochaete colonies. This requires the assembly of a set of genetic rules that constitute the organism’s genome, and which govern the dynamic behaviour of the colonies. Using a simple set of assumptions, partly based on the empirical rules for cell division formulated in the 1800’s, we have realistically modeled the behaviour of colonies during undisturbed growth, after damage of the meristematic layer, and on the collision of adjacent colonies. In addition, we can successfully extract a description of a Coleochaete colony from a microscope image, and feed this description into the computer modelling software. This provides an important and crucial link between biological experiments and computer modelling.