Tim Rudge

Researchgate.net/profile/Tim_Rudge/
Click here to download CV (PDF, 112K)
CellModeller software
Research

CellModeller:Biofilm
With PJ Steiner, I have developed a large-scale biophysical simulation method for bacterial biofilms. Using GPUs this method can generate >100K cells in a 2 or 3 hours starting from a single founder cell. Biophysical effects are important in generating morphology, which is one of my targets for engineering. The biophysical model is integrated into CellModeller, which is a framework we have developed for combined physical and genetic modelling of cellular systems. CellModeller is an open source package and code is available at Github.
I am using this system in combination with confocal microscopy to study the interaction between genetic pattern formation and biophysical shape generation. This work was done in collaboration with PJ Steiner.
I am using this system in combination with confocal microscopy to study the interaction between genetic pattern formation and biophysical shape generation. This work was done in collaboration with PJ Steiner.
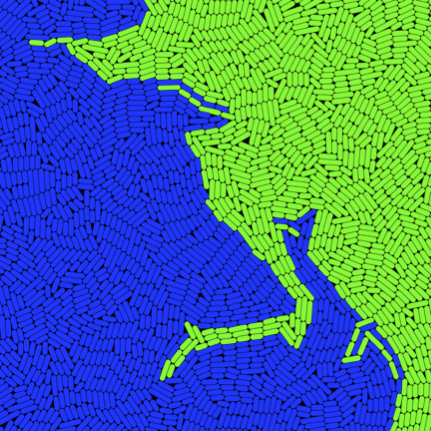
Emergent spatial organization of biofilms
Biofilms play an important role in pathogenesis and have potential uses in biotechnology. Understanding spatial organization of biofilms is critical for attempts to either engineer or disrupt them. We are studying spatial patterns in E. coli biofilms, and developing approaches to modulate and control these patterns. Combining CellModeller:Biofilm with confocal microscopy, I can compare large populations grown from similar distributions of founding cells. The results suggest that the large-scale morphology of these patterns is primarily an emergent property of local physical interactions between cells. Simple growth behaviour, and physical interactions are sufficient to reproduce complex patterns with striking similarity. This work is being done in collaboration with Fernan Federici and PJ Steiner.
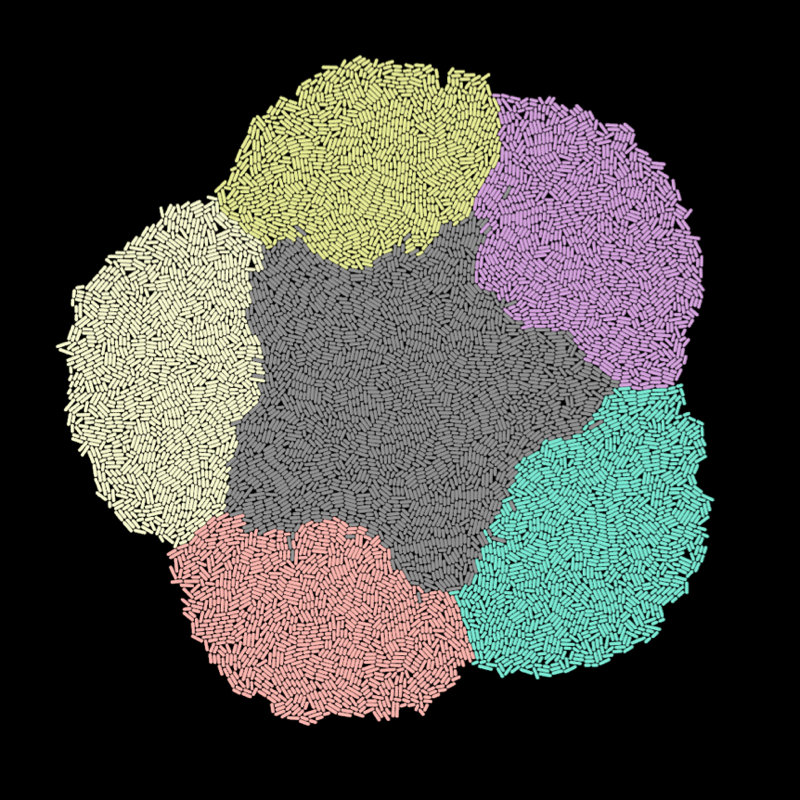
Engineering biofilm morphology
I am developing genetic approaches to modulating cell growth and division, the key drivers of morphogenesis. In combination with pattern formation mechanisms, these growth effectors will be used to generate form in E. coli colonies.
CellModeller:Biofilm provides a way to explore the possible morphogenetic systems, incorporating genetics, signalling, and cell growth and division. The parallel GPU algorithm combined with our GPU cluster mean that we can perform high-throughput computational screens.
CellModeller:Biofilm provides a way to explore the possible morphogenetic systems, incorporating genetics, signalling, and cell growth and division. The parallel GPU algorithm combined with our GPU cluster mean that we can perform high-throughput computational screens.

CellModeller:Plant
Based on my previous work, and that of Lionel Dupuy, I have implemented parallel GPU plant tissue biophysical models in 2D that can simulate growth to 10,000 cell stage in less than a minute. These are integrated into the CellModeller framework, allowing me to explore designs for morphogenetic systems in plants. Working with Fernan Federici, I am studying possible uses of plant genetic regulatory elements, and growth effectors for engineering plant morphology. I am also working with Nuri Purswani, who is collecting confocal timelapse data of Marchantia polymorpha. We are analysing cell growth and division, and using this to build dynamic models of Marchantia morphogenesis.
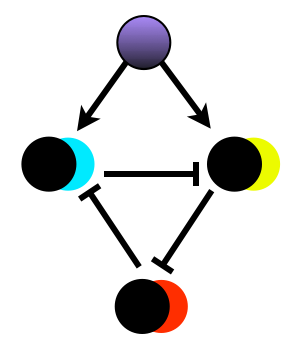
Synthetic segmentation
An important mechanism in development is response to spatial signals to generate domains of gene expression. Working with Fernan Federici, I am using computational modelling to enumerate possible 3-node repressor networks that respond to gradients of AHL inducer. This modelling is based on relative characterisation of promoter transcription rates, using two-channel fluorescence measurements. I am using high-throughput GPU-based methods to assess the ability of each network to create and maintain domains under noisy conditions. We are building and assaying networks predicted to perform well, and those predicted to perform poorly. This will allow us to validate the modelling approach, and create modules for spatial pattern formation. This work is being done in collaboration with Fernan Federici, and Microsoft Research Cambridge.
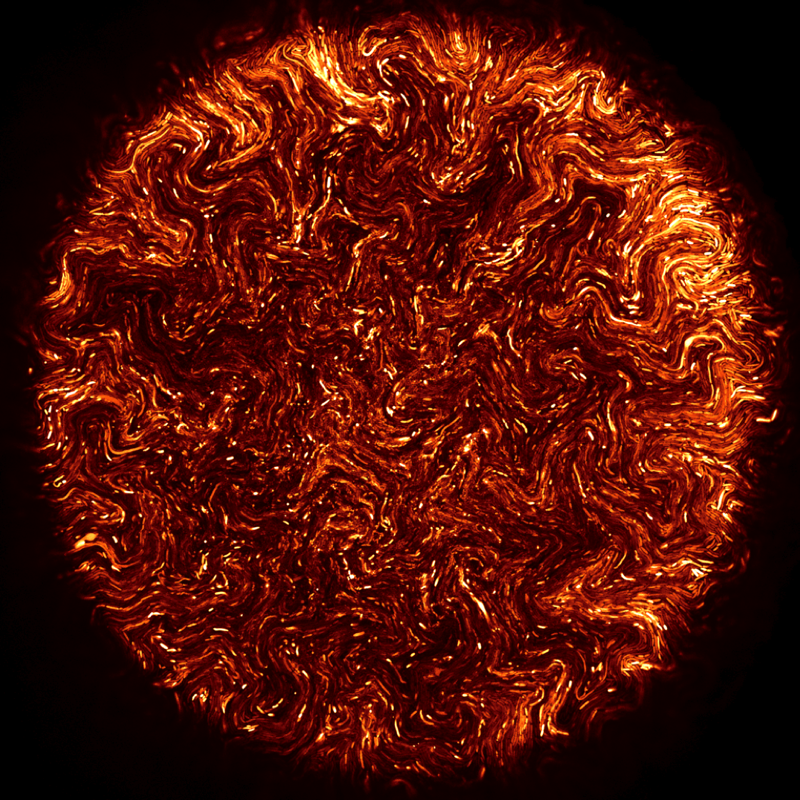
Confocal microscopy
I am using confocal microscopy to study microcolony and biofilm development at single cell level, in 3-dimensions, and over time courses. Some of these images were included in a recent Cell Picture Show on biofilms. Fernan Federici, PJ Steiner and I were also awarded a Wellcome Trust Image award in 2012
We are also working with architect David Benjamin (http://www.thelivingnewyork.com, http://www.arch.columbia.edu/labs/living-architecture-lab) to use CellModeller to develop a conceptual workflow for biomaterials design, in which spatially patterned gene expression leads to deposition of materials with different structural properties.
We are also working with architect David Benjamin (http://www.thelivingnewyork.com, http://www.arch.columbia.edu/labs/living-architecture-lab) to use CellModeller to develop a conceptual workflow for biomaterials design, in which spatially patterned gene expression leads to deposition of materials with different structural properties.
Research Experience
2010 - present: Microsoft PhD studentship, Plant Sciences, University of Cambridge, UK
2009 – 2010: Research Associate, Centre for Medical Image Computing, Dept. of Computer Science, University College London
2007 – 2008: Quantitative Developer, Endeavour Capital Management LLP, London
2006 – 2007: Risk Analyst, InterGen (Australia), Brisbane
2006 - 2007: Graduate Certificate in Business, Queensland University of Technology, Brisbane, Australia
2006 – 2006: Software Engineer, Leica Geosystems, Brisbane
2004 – 2006: Research Officer, University of Queensland, ARC Centre for Complex Systems, Advanced Computational Modelling Centre
2002 – 2004: Research Associate, Cambridge University, Plant Sciences
2001 - 2002: Masters by Research (Computer Science), University College London , London, UK
1999 – 2001: EFL Instructor, Tokyo, Japan
1995 - 1998: BEng(Hons) (Mechatronics), Leeds University (Mechanical/Electrical Engineering), Leeds, UK
2010 - present: Microsoft PhD studentship, Plant Sciences, University of Cambridge, UK
2009 – 2010: Research Associate, Centre for Medical Image Computing, Dept. of Computer Science, University College London
2007 – 2008: Quantitative Developer, Endeavour Capital Management LLP, London
2006 – 2007: Risk Analyst, InterGen (Australia), Brisbane
2006 - 2007: Graduate Certificate in Business, Queensland University of Technology, Brisbane, Australia
2006 – 2006: Software Engineer, Leica Geosystems, Brisbane
2004 – 2006: Research Officer, University of Queensland, ARC Centre for Complex Systems, Advanced Computational Modelling Centre
2002 – 2004: Research Associate, Cambridge University, Plant Sciences
2001 - 2002: Masters by Research (Computer Science), University College London , London, UK
1999 – 2001: EFL Instructor, Tokyo, Japan
1995 - 1998: BEng(Hons) (Mechatronics), Leeds University (Mechanical/Electrical Engineering), Leeds, UK
Publications
Rudge*, T., Federici*, F., Steiner*, P., Kan, A., Haseloff, J., Cell shape-driven instability generates self-organised, fractal patterning of cell layers, ACS Synthetic Biology, May 2013
Federici F., Rudge T., Pollak B., Haseloff J., Gutierrez R., Synthetic Biology: opportunities for Chilean bioindustry and education, Biological Research, 46:383-393, June 2013
Correia, T., Rudge, T., Koch, M., Ntziachristos, V., Arridge, S., Wavelet-based data and solution compression for efficient image reconstruction in fluorescence diffuse optical tomography, J. Biomed. Optics,18(8):86008, 2013
Correia, T., Rudge, T., Arridge, S., Efficient image reconstruction in fluorescence diffuse optical tomography (fDOT) using data and solution compression, Proc. SPIE, 8799, 2013
Rudge*, T., Steiner*, P., Phillips, A., Haseloff, J., Computational modeling of synthetic microbial biofilms, ACS Synthetic Biology, 1(8):345-352, 2012
Ducros, N., D'andrea, C., Valentini, G., Rudge, T., Arridge, S., Bassi, A., Full-wavelet approach for fluorescence diffuse optical tomography with structured illumination, Optics Letters, 35(21):3676-3678, 2010
Rudge, T., Soloviev, V., Arridge, S., Fast image reconstruction in fluoresence optical tomography using data compression, Optics Letters, 35(5):763-765, 2010
Rudge, T. and Burrage, K., Effects of Intrinsic and Extrinsic Noise Can Accelerate Juxtacrine Pattern Formation, Bulletin of Mathematical Biology, 70(4):971-991, 2008
Dupuy, L., Mackenzie, J., Rudge, T., Haseloff, J., A System for Modelling Cell-Cell Interactions during Plant Morphogenesis, Annals of Botany, 101(8):1255-1265, 2008
Rudge, T. and Geard, N., Evolving Gene Regulatory Networks for Cellular Morphogenesis, Advances in Natural Computation, 3:239-252, 2005
Rudge, T. and Haseloff, J., A Computational Model of Cellular Morphogenesis in Plants, Lecture Notes in Computer Science, 3630:78-87, 2005
Rudge*, T., Federici*, F., Steiner*, P., Kan, A., Haseloff, J., Cell shape-driven instability generates self-organised, fractal patterning of cell layers, ACS Synthetic Biology, May 2013
Federici F., Rudge T., Pollak B., Haseloff J., Gutierrez R., Synthetic Biology: opportunities for Chilean bioindustry and education, Biological Research, 46:383-393, June 2013
Correia, T., Rudge, T., Koch, M., Ntziachristos, V., Arridge, S., Wavelet-based data and solution compression for efficient image reconstruction in fluorescence diffuse optical tomography, J. Biomed. Optics,18(8):86008, 2013
Correia, T., Rudge, T., Arridge, S., Efficient image reconstruction in fluorescence diffuse optical tomography (fDOT) using data and solution compression, Proc. SPIE, 8799, 2013
Rudge*, T., Steiner*, P., Phillips, A., Haseloff, J., Computational modeling of synthetic microbial biofilms, ACS Synthetic Biology, 1(8):345-352, 2012
Ducros, N., D'andrea, C., Valentini, G., Rudge, T., Arridge, S., Bassi, A., Full-wavelet approach for fluorescence diffuse optical tomography with structured illumination, Optics Letters, 35(21):3676-3678, 2010
Rudge, T., Soloviev, V., Arridge, S., Fast image reconstruction in fluoresence optical tomography using data compression, Optics Letters, 35(5):763-765, 2010
Rudge, T. and Burrage, K., Effects of Intrinsic and Extrinsic Noise Can Accelerate Juxtacrine Pattern Formation, Bulletin of Mathematical Biology, 70(4):971-991, 2008
Dupuy, L., Mackenzie, J., Rudge, T., Haseloff, J., A System for Modelling Cell-Cell Interactions during Plant Morphogenesis, Annals of Botany, 101(8):1255-1265, 2008
Rudge, T. and Geard, N., Evolving Gene Regulatory Networks for Cellular Morphogenesis, Advances in Natural Computation, 3:239-252, 2005
Rudge, T. and Haseloff, J., A Computational Model of Cellular Morphogenesis in Plants, Lecture Notes in Computer Science, 3630:78-87, 2005